使用Transformer进行抄袭检测
点击下方卡片,关注“新机器视觉”公众号
视觉/图像重磅干货,第一时间送达
来源:磐创AI
动机
在许多行业中,尤其是在学术界,抄袭是一个重大问题。随着互联网和开放信息的兴起,这种现象甚至变得更加严重,任何人都可以通过点击访问特定主题的任何信息。
基于这一观察,研究人员一直在尝试使用不同的文本分析方法解决这个问题。在这篇概念文章中,我们将尝试解决抄袭检测工具的两个主要限制:(1)内容改写抄袭和(2)内容翻译抄袭。
(1) 对于传统工具来说,重新表述的内容可能很难捕捉到,因为它们没有考虑整体上下文的同义词和反义词。
(2) 使用与原文不同语言编写的内容也是一个巨大的问题,即使是最先进的基于机器学习的工具也面临着这个问题,因为上下文完全转移到了另一种语言。
在这篇概念性的博客文章中,我们将解释如何使用基于Transformer的模型以创新的方式解决这两个挑战。
首先,我们将带你了解分析方法,描述从数据收集到性能分析的整个工作流程。然后,我们将深入探讨解决方案的科学/技术实现,然后展示最终结果。
问题陈述
假设你有兴趣构建一个学术内容管理平台。你可能希望只接受在你的平台上没有共享过的文章。在这种情况下,你的目标将是拒绝所有与现有文章相似度超过某个阈值的新文章。
为了说明这种情况,我们将使用cord-19数据集,这是由Allen Institute for AI在Kaggle上免费提供的开放研究挑战数据集。
https://allenai.org/
分析方法
在进一步进行分析之前,让我们从以下问题明确我们在这里试图实现的目标:
问题:我们能否在我们的数据库中找到一个或多个与新提交的文档相似(超过某个阈值)的文档?
下面的工作流程突出显示了回答这个问题所需的所有主要步骤。
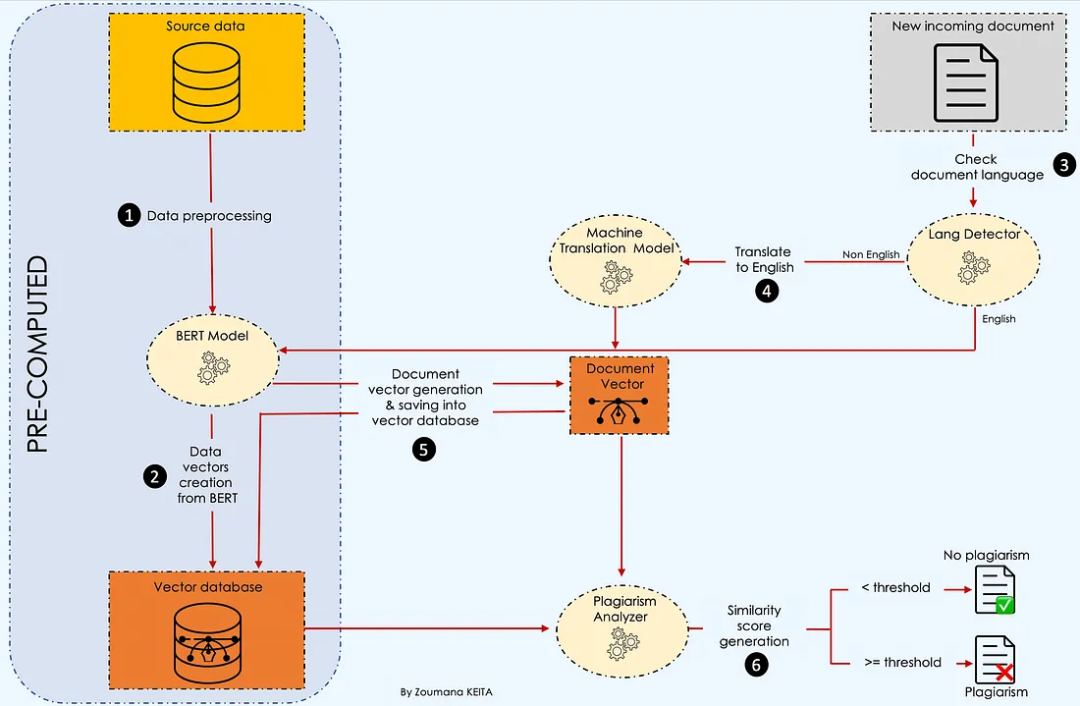
让我们了解这里正在发生的事情 💡。
在收集源数据后,我们首先对内容进行预处理,然后使用BERT创建一个向量数据库。
然后,每当我们有一个新的文档进入时,我们检查语言并进行抄袭检测。更多详细信息将在文章后面给出。
科学实施
本节专注于分析方法中各个部分的技术实施。
数据预处理
我们只对源数据的摘要列感兴趣,为了简单起见,我们将仅使用100个观察结果来加快预处理的速度。
import pandas as pddef preprocess_data(data_path, sample_size):# Read the data from specific pathdata = pd.read_csv(data_path, low_memory=False)# Drop articles without Abstractdata = data.dropna(subset = ['abstract']).reset_index(drop = True)# Get "sample_size" random articlesdata = data.sample(sample_size)[['abstract']]return data# Read data & preprocess itdata_path = "./data/cord19_source_data.csv"source_data = preprocess_data(data_path, 100)
以下是源数据集的五个随机观察结果。

文档向量化器
在引言中观察到的挑战分别导致选择以下两个基于Transformer的模型:
(1) BERT模型:用于解决第一个限制,因为它提供了文本信息更好的上下文表示。为此,我们将使用以下功能:
create_vector_from_text:用于生成单个文档的向量表示。
create_vector_database:负责创建一个数据库,其中包含每个文档的相应向量。
# Useful librariesimport numpy as npimport torchfrom keras.preprocessing.sequence import pad_sequencesfrom transformers import BertTokenizer, AutoModelForSequenceClassification# Load bert modelmodel_path = "bert-base-uncased"tokenizer = BertTokenizer.from_pretrained(model_path,do_lower_case=True)model = AutoModelForSequenceClassification.from_pretrained(model_path,output_attentions=False,output_hidden_states=True)def create_vector_from_text(tokenizer, model, text, MAX_LEN = 510):input_ids = tokenizer.encode(text,add_special_tokens = True,max_length = MAX_LEN,)results = pad_sequences([input_ids], maxlen=MAX_LEN, dtype="long",truncating="post", padding="post")# Remove the outer list.input_ids = results[0]# Create attention masksattention_mask = [int(i>0) for i in input_ids]# Convert to tensors.input_ids = torch.tensor(input_ids)attention_mask = torch.tensor(attention_mask)# Add an extra dimension for the "batch" (even though there is only one# input in this batch.)input_ids = input_ids.unsqueeze(0)attention_mask = attention_mask.unsqueeze(0)# Put the model in "evaluation" mode, meaning feed-forward operation.model.eval()# Run the text through BERT, and collect all of the hidden states produced# from all 12 layers.with torch.no_grad():logits, encoded_layers = model(input_ids = input_ids,token_type_ids = None,attention_mask = attention_mask,return_dict=False)layer_i = 12 # The last BERT layer before the classifier.batch_i = 0 # Only one input in the batch.token_i = 0 # The first token, corresponding to [CLS]# Extract the vector.vector = encoded_layers[layer_i][batch_i][token_i]# Move to the CPU and convert to numpy ndarray.vector = vector.detach().cpu().numpy()return(vector)def create_vector_database(data):# The list of all the vectorsvectors = []# Get overall text datasource_data = data.abstract.values# Loop over all the comment and get the embeddingsfor text in tqdm(source_data):# Get the embeddingvector = create_vector_from_text(tokenizer, model, text)#add it to the listvectors.append(vector)data["vectors"] = vectorsdata["vectors"] = data["vectors"].apply(lambda emb: np.array(emb))data["vectors"] = data["vectors"].apply(lambda emb: emb.reshape(1, -1))return data# Create the vector databasevector_database = create_vector_database(source_data)vector_database.sample(5)
第94行显示了向量数据库中的五个随机观察结果,包括新向量列。

(2) 使用机器翻译Transformer模型将传入文档的语言翻译为英语,因为我们的源文档是英文的。只有当文档的语言是以下五种语言之一时,才执行翻译:德语、法语、日语、希腊语和俄语。以下是使用MarianMT模型实现此逻辑的辅助函数。
from langdetect import detect, DetectorFactoryDetectorFactory.seed = 0def translate_text(text, text_lang, target_lang='en'):# Get the name of the modelmodel_name = f"Helsinki-NLP/opus-mt-{text_lang}-{target_lang}"# Get the tokenizertokenizer = MarianTokenizer.from_pretrained(model_name)# Instantiate the modelmodel = MarianMTModel.from_pretrained(model_name)# Translation of the textformated_text = ">>{}<< {}".format(text_lang, text)translation = model.generate(**tokenizer([formated_text], return_tensors="pt", padding=True))translated_text = [tokenizer.decode(t, skip_special_tokens=True) for t in translation][0]return translated_text
抄袭分析器
当传入文档的向量与数据库中的某个向量在一定阈值水平上相似时,就存在抄袭。
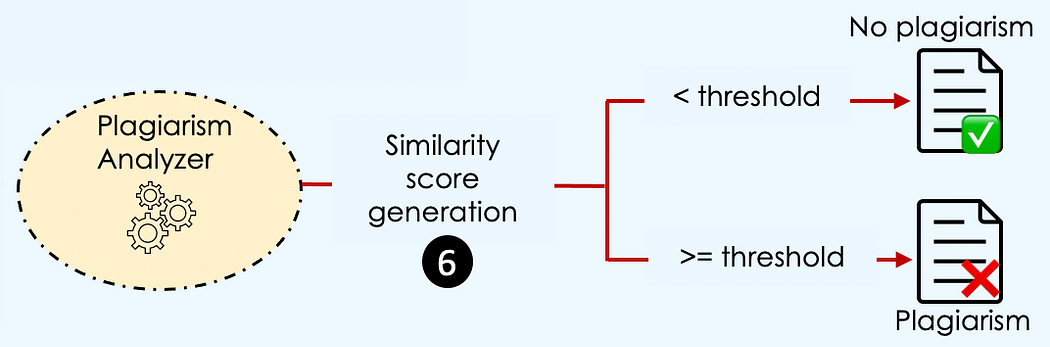
但是,什么时候两个向量是相似的?→ 当它们具有相同的大小和方向时。
这个定义要求我们的向量具有相同的大小,这可能是一个问题,因为文档向量的维度取决于该文档的长度。幸运的是,我们有多种相似度测量方法可以用来解决这个问题,其中之一就是余弦相似度,我们将在本例中使用它。
如果你对其他方法感兴趣,可以参考James Briggs的这篇精彩内容。他解释了每种方法的工作原理、优点,并指导你如何实施它们。
https://www.pinecone.io/learn/semantic-search/
抄袭分析是使用run_plagiarism_analysis函数执行的。我们首先使用check_incoming_document函数检查文档语言,必要时执行正确的翻译。
最终结果是一个包含四个主要值的字典:
similarity_score:传入文章与数据库中最相似的现有文章之间的得分。
is_plagiarism:如果相似度得分等于或超过阈值,则值为true。否则为false。
most_similar_article:最相似文章的文本信息。
article_submitted:提交审批的文章。
def process_document(text):"""Create a vector for given text and adjust it for cosine similarity search"""text_vect = create_vector_from_text(tokenizer, model, text)text_vect = np.array(text_vect)text_vect = text_vect.reshape(1, -1)return text_vectdef is_plagiarism(similarity_score, plagiarism_threshold):return similarity_score < plagiarism_thresholddef check_incoming_document(incoming_document):text_lang = detect(incoming_document)language_list = ['de', 'fr', 'el', 'ja', 'ru']final_result = ""if(text_lang == 'en'):final_result = incoming_documentelif(text_lang not in language_list):final_result = Noneelse:# Translate in Englishfinal_result = translate_text(incoming_document, text_lang)return final_resultdef run_plagiarism_analysis(query_text, data, plagiarism_threshold=0.8):top_N=3# Check the language of the query/incoming text and translate if required.document_translation = check_incoming_document(query_text)if(document_translation is None):print("Only the following languages are supported: English, French, Russian, German, Greek and Japanese")exit(-1)else:# Preprocess the document to get the required vector for similarity analysisquery_vect = process_document(document_translation)# Run similarity Searchdata["similarity"] = data["vectors"].apply(lambda x: cosine_similarity(query_vect, x))data["similarity"] = data["similarity"].apply(lambda x: x[0][0])similar_articles = data.sort_values(by='similarity', ascending=False)[1:top_N+1]formated_result = similar_articles[["abstract", "paper_id", "similarity"]].reset_index(drop = True)similarity_score = formated_result.iloc[0]["similarity"]most_similar_article = formated_result.iloc[0]["abstract"]is_plagiarism_bool = is_plagiarism(similarity_score, plagiarism_threshold)plagiarism_decision = {'similarity_score': similarity_score,'is_plagiarism': is_plagiarism_bool,'most_similar_article': most_similar_article,'article_submitted': query_text}return plagiarism_decision
系统实验
我们已经涵盖并实施了工作流程的所有组件。现在,是时候使用我们的系统来测试三种被系统接受的语言:德语、法语、日语、希腊语和俄语。
评估
以下是我们要检查作者是否抄袭的文章摘要文本。
英文文章
这篇文章实际上是源数据中的一个示例。
english_article_to_check = "The need for multidisciplinary research to address today's complex health and environmental challenges has never been greater. The One Health (OH) approach to research ensures that human, animal, and environmental health questions are evaluated in an integrated and holistic manner to provide a more comprehensive understanding of the problem and potential solutions than would be possible with siloed approaches. However, the OH approach is complex, and there is limited guidance available for investigators regarding the practical design and implementation of OH research. In this paper we provide a framework to guide researchers through conceptualizing and planning an OH study. We discuss key steps in designing an OH study, including conceptualization of hypotheses and study aims, identification of collaborators for a multi-disciplinary research team, study design options, data sources and collection methods, and analytical methods. We illustrate these concepts through the presentation of a case study of health impacts associated with land application of biosolids. Finally, we discuss opportunities for applying an OH approach to identify solutions to current global health issues, and the need for cross-disciplinary funding sources to foster an OH approach to research."# Select an existing article from the databasenew_incoming_text = source_data.iloc[0]['abstract']# Run the plagiarism detectionanalysis_result = run_plagiarism_analysis(new_incoming_text, vector_database, plagiarism_threshold=0.8)
运行系统后,我们得到了一个相似度得分为1,与现有文章完全匹配。这是显而易见的,因为我们从数据库中取了完全相同的文章。
法文文章
这篇文章可以从法国农业网站免费获取。
french_article_to_check = """Les Réseaux d’Innovation et de Transfert Agricole (RITA) ont été créés en 2011 pour mieux connecter la recherche et le développement agricole, intra et inter-DOM, avec un objectif d’accompagnement de la diversification des productions locales. Le CGAAER a été chargé d'analyser ce dispositif et de proposer des pistes d'action pour améliorer la chaine Recherche – Formation – Innovation – Développement – Transfert dans les outre-mer dans un contexte d'agriculture durable, au profit de l'accroissement de l'autonomie alimentaire."""
analysis_result = run_plagiarism_analysis(french_article_to_check, vector_database, plagiarism_threshold=0.8)analysis_result

在这种情况下,没有发生抄袭,因为相似度得分低于阈值。
德文文章
假设有人非常喜欢数据库中的第五篇文章,并决定将其翻译成德语。现在让我们看看系统如何判断这篇文章。
german_article_to_check = """Derzeit ist eine Reihe strukturell und funktionell unterschiedlicher temperaturempfindlicher Elemente wie RNA-Thermometer bekannt, die eine Vielzahl biologischer Prozesse in Bakterien, einschließlich der Virulenz, steuern. Auf der Grundlage einer Computer- und thermodynamischen Analyse der vollständig sequenzierten Genome von 25 Salmonella enterica-Isolaten wurden ein Algorithmus und Kriterien für die Suche nach potenziellen RNA-Thermometern entwickelt. Er wird es ermöglichen, die Suche nach potentiellen Riboschaltern im Genom anderer gesellschaftlich wichtiger Krankheitserreger durchzuführen. Für S. enterica wurden neben dem bekannten 4U-RNA-Thermometer vier Hairpin-Loop-Strukturen identifiziert, die wahrscheinlich als weitere RNA-Thermometer fungieren. Sie erfüllen die notwendigen und hinreichenden Bedingungen für die Bildung von RNA-Thermometern und sind hochkonservative nichtkanonische Strukturen, da diese hochkonservativen Strukturen im Genom aller 25 Isolate von S. enterica gefunden wurden. Die Hairpins, die eine kreuzförmige Struktur in der supergewickelten pUC8-DNA bilden, wurden mit Hilfe der Rasterkraftmikroskopie sichtbar gemacht."""
analysis_result = run_plagiarism_analysis(german_article_to_check, vector_database, plagiarism_threshold=0.8)analysis_result

相似度达到了97% - 模型捕捉到了这一点!结果非常令人印象深刻。这篇文章绝对是一个剽窃作品。
结论
恭喜!现在你拥有了构建更强大的抄袭检测系统所需的所有工具,使用BERT和机器翻译模型结合余弦相似度。
感谢阅读!
其他资源
https://huggingface.co/docs/transformers/model_doc/marian
https://github.com/keitazoumana/Medium-Articles-Notebooks/blob/main/Plagiarism_detection.ipynb
https://allenai.org/
声明:部分内容来源于网络,仅供读者学习、交流之目的。文章版权归原作者所有。如有不妥,请联系删除。
—THE END—
评论