UNet 和 UNet++:医学影像经典分割网络对比

来源:极市平台 本文约3000字,建议阅读5分钟
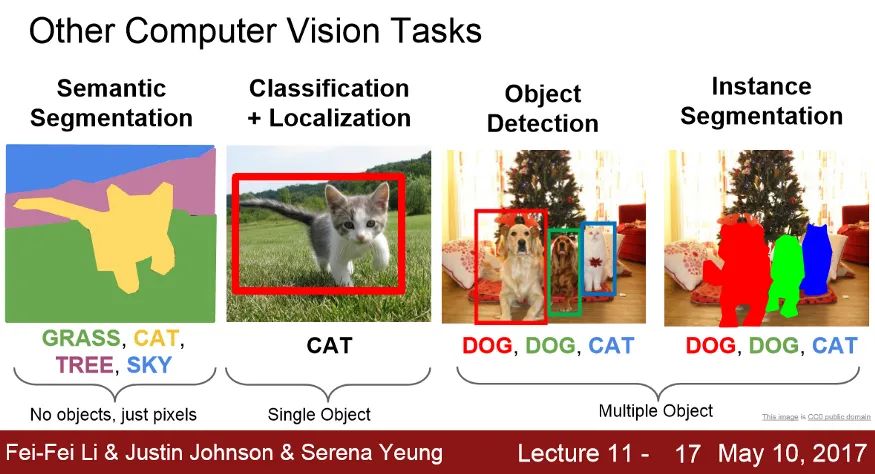
本文介绍了医学影像经典分割网络的对比。
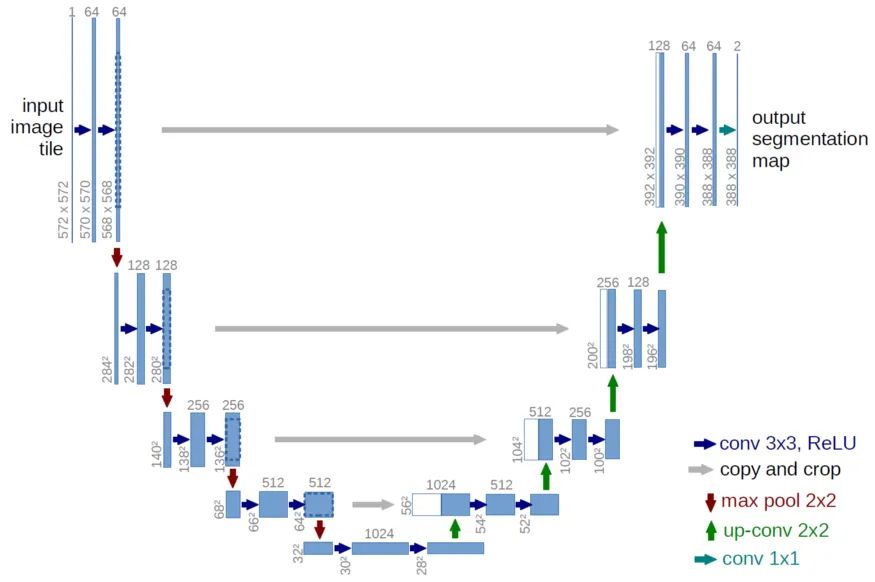
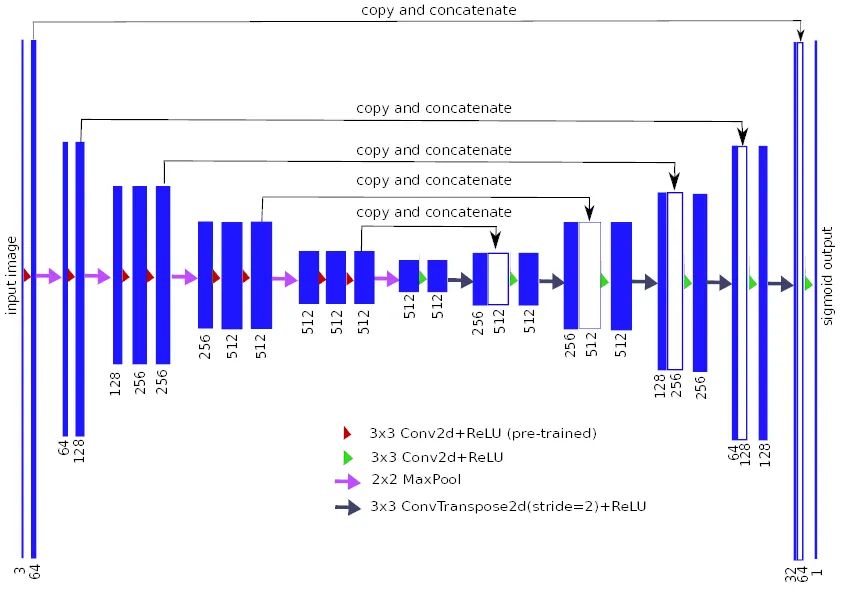
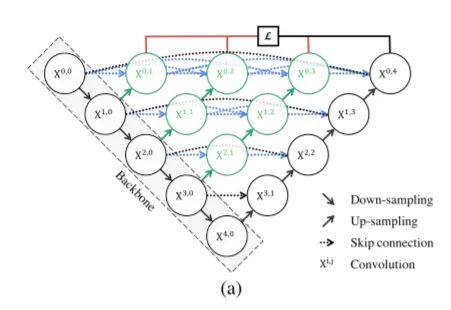
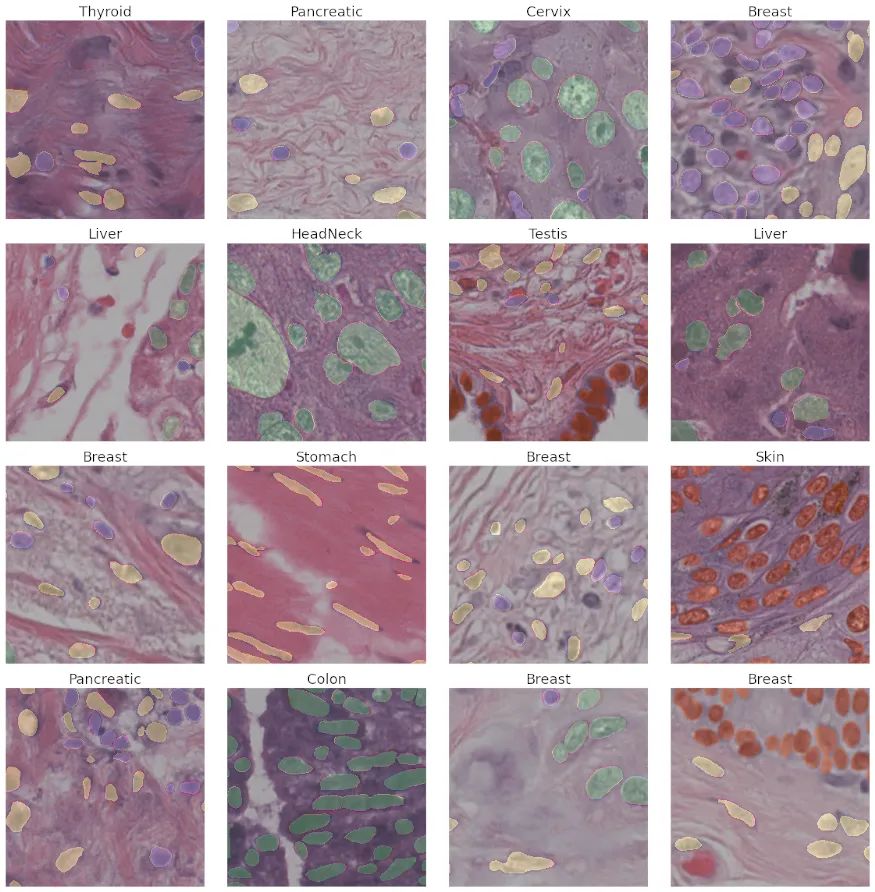
根据论文, Unet++的表现似乎优于原来的Unet。就像在Unet中一样,这里可以使用多个编码器(骨干)来为输入图像生成强特征。
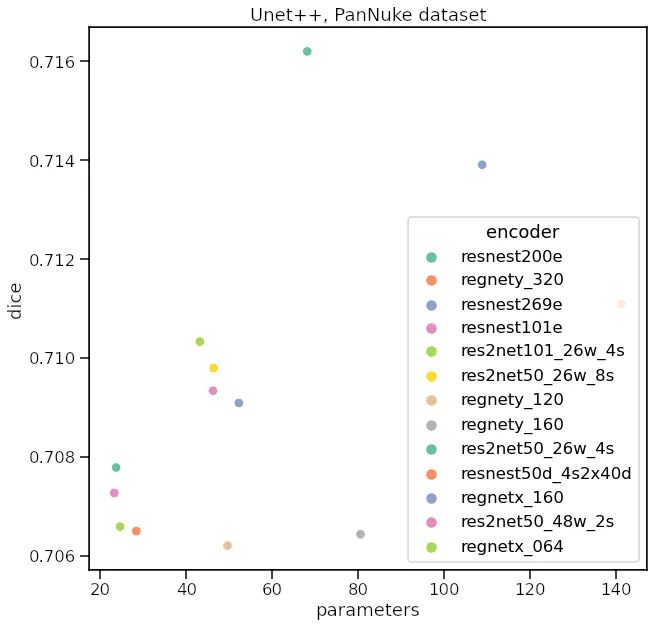
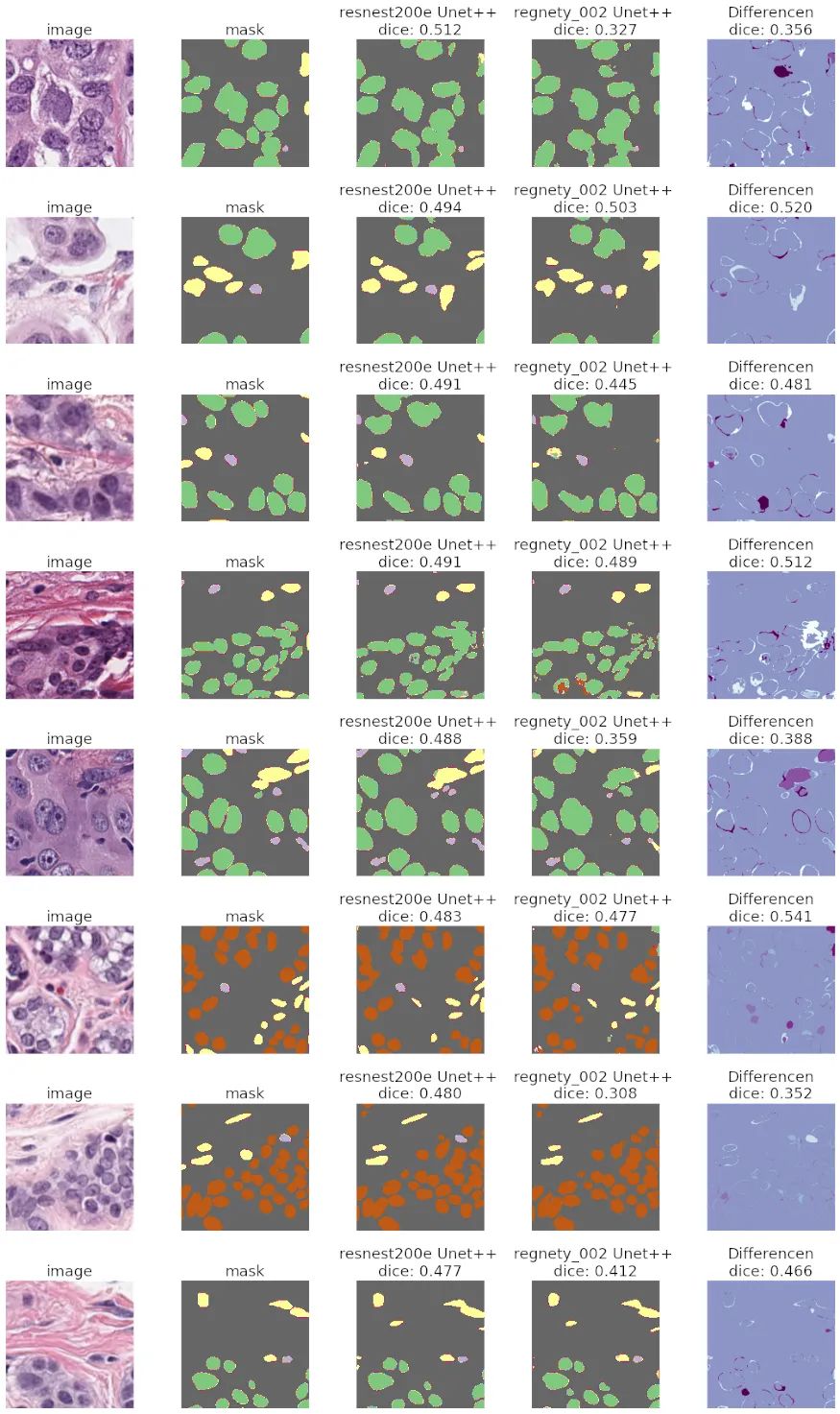
我应该使用哪个编码器?
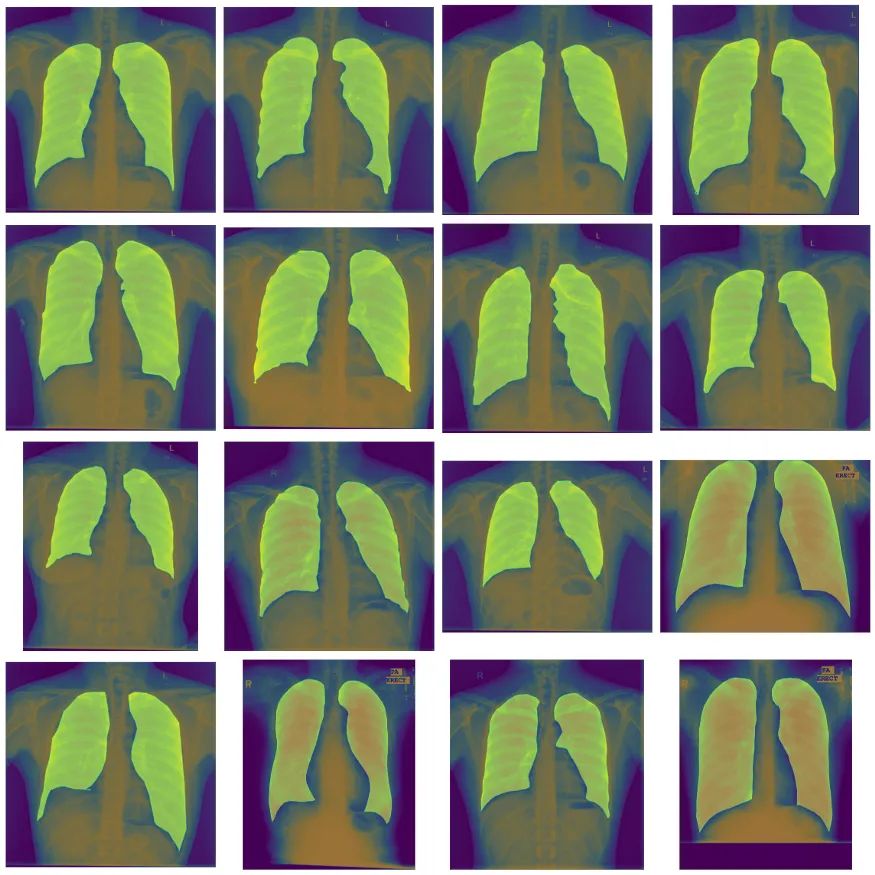
定义数据集和增强。我们将调整图像大小为256×256,并对训练数据集应用一些大的增强。
import albumentations as A
from torch.utils.data import Dataset, DataLoader
from collections import OrderedDict
class ChestXRayDataset(Dataset):
def __init__(
self,
images,
masks,
transforms):
self.images = images
self.masks = masks
self.transforms = transforms
def __len__(self):
return(len(self.images))
def __getitem__(self, idx):
"""Will load the mask, get random coordinates around/with the mask,
load the image by coordinates
"""
sample_image = imread(self.images[idx])
if len(sample_image.shape) == 3:
sample_image = sample_image[..., 0]
sample_image = np.expand_dims(sample_image, 2) / 255
sample_mask = imread(self.masks[idx]) / 255
if len(sample_mask.shape) == 3:
sample_mask = sample_mask[..., 0]
augmented = self.transforms(image=sample_image, mask=sample_mask)
sample_image = augmented['image']
sample_mask = augmented['mask']
sample_image = sample_image.transpose(2, 0, 1) # channels first
sample_mask = np.expand_dims(sample_mask, 0)
data = {'features': torch.from_numpy(sample_image.copy()).float(),
'mask': torch.from_numpy(sample_mask.copy()).float()}
return(data)
def get_valid_transforms(crop_size=256):
return A.Compose(
[
A.Resize(crop_size, crop_size),
],
p=1.0)
def light_training_transforms(crop_size=256):
return A.Compose([
A.RandomResizedCrop(height=crop_size, width=crop_size),
A.OneOf(
[
A.Transpose(),
A.VerticalFlip(),
A.HorizontalFlip(),
A.RandomRotate90(),
A.NoOp()
], p=1.0),
])
def medium_training_transforms(crop_size=256):
return A.Compose([
A.RandomResizedCrop(height=crop_size, width=crop_size),
A.OneOf(
[
A.Transpose(),
A.VerticalFlip(),
A.HorizontalFlip(),
A.RandomRotate90(),
A.NoOp()
], p=1.0),
A.OneOf(
[
A.CoarseDropout(max_holes=16, max_height=16, max_width=16),
A.NoOp()
], p=1.0),
])
def heavy_training_transforms(crop_size=256):
return A.Compose([
A.RandomResizedCrop(height=crop_size, width=crop_size),
A.OneOf(
[
A.Transpose(),
A.VerticalFlip(),
A.HorizontalFlip(),
A.RandomRotate90(),
A.NoOp()
], p=1.0),
A.ShiftScaleRotate(p=0.75),
A.OneOf(
[
A.CoarseDropout(max_holes=16, max_height=16, max_width=16),
A.NoOp()
], p=1.0),
])
def get_training_trasnforms(transforms_type):
if transforms_type == 'light':
return(light_training_transforms())
elif transforms_type == 'medium':
return(medium_training_transforms())
elif transforms_type == 'heavy':
return(heavy_training_transforms())
else:
raise NotImplementedError("Not implemented transformation configuration")
定义模型和损失函数。这里我们使用带有regnety\_004编码器的Unet++,并使用RAdam + Lookahed优化器使用DICE + BCE损失之和进行训练。
import torch
import segmentation_models_pytorch as smp
import numpy as np
import matplotlib.pyplot as plt
from catalyst import dl, metrics, core, contrib, utils
import torch.nn as nn
from skimage.io import imread
import os
from sklearn.model_selection import train_test_split
from catalyst.dl import CriterionCallback, MetricAggregationCallback
encoder = 'timm-regnety_004'
model = smp.UnetPlusPlus(encoder, classes=1, in_channels=1)
#model.cuda()
learning_rate = 5e-3
encoder_learning_rate = 5e-3 / 10
layerwise_params = {"encoder*": dict(lr=encoder_learning_rate, weight_decay=0.00003)}
model_params = utils.process_model_params(model, layerwise_params=layerwise_params)
base_optimizer = contrib.nn.RAdam(model_params, lr=learning_rate, weight_decay=0.0003)
optimizer = contrib.nn.Lookahead(base_optimizer)
scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, factor=0.25, patience=10)
criterion = {
"dice": DiceLoss(mode='binary'),
"bce": nn.BCEWithLogitsLoss()
}
定义回调函数并训练!
callbacks = [
# Each criterion is calculated separately.
CriterionCallback(
input_key="mask",
prefix="loss_dice",
criterion_key="dice"
),
CriterionCallback(
input_key="mask",
prefix="loss_bce",
criterion_key="bce"
),
# And only then we aggregate everything into one loss.
MetricAggregationCallback(
prefix="loss",
mode="weighted_sum",
metrics={
"loss_dice": 1.0,
"loss_bce": 0.8
},
),
# metrics
IoUMetricsCallback(
mode='binary',
input_key='mask',
)
]
runner = dl.SupervisedRunner(input_key="features", input_target_key="mask")
runner.train(
model=model,
criterion=criterion,
optimizer=optimizer,
scheduler=scheduler,
loaders=loaders,
callbacks=callbacks,
logdir='../logs/xray_test_log',
num_epochs=100,
main_metric="loss",
minimize_metric=True,
verbose=True,
)
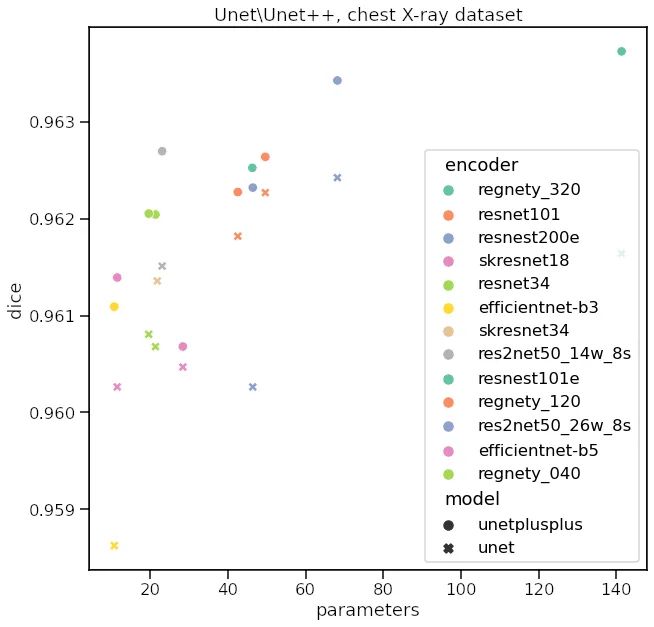
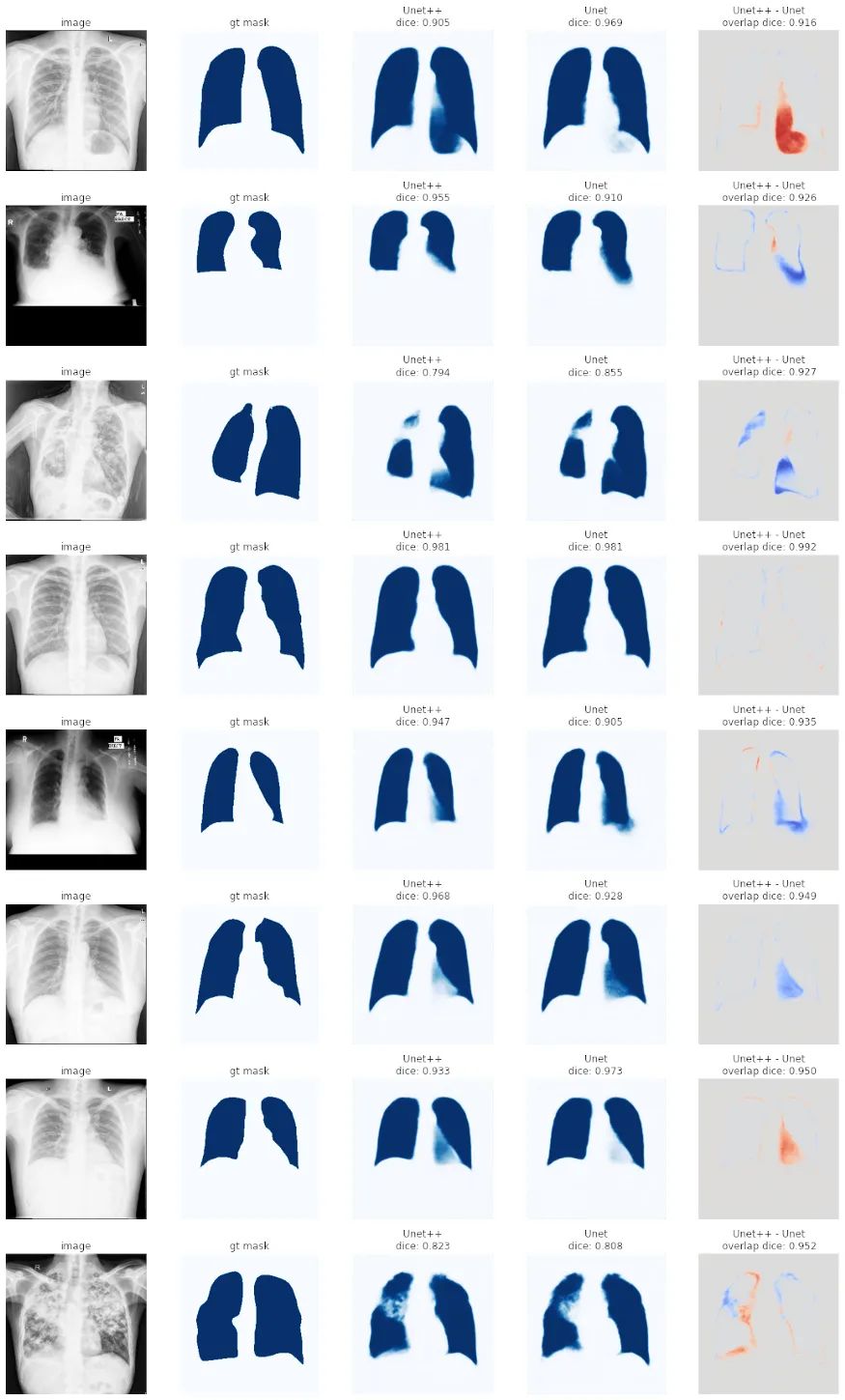
总结
英文原文:
https://towardsdatascience.com/the-best-approach-to-semantic-segmentation-of-biomedical-images-bbe4fd78733f
评论