2022年CVPR医学影像论文合集(不完全收集)
“ 2022年的CVPR也有一些关于医学影像的文章,这里整理了目前发现的一些相关论文以及开源代码,方便大家查看。”
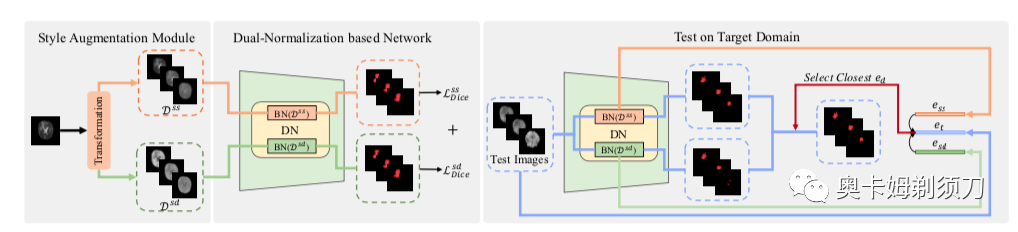
[1] Generalizable Cross-modality Medical Image Segmentation via Style Augmentation and Dual Normalization(通过风格增强和双重归一化的可泛化跨模态医学图像分割)
For medical image segmentation, imagine if a model was only trained using MR images in source domain, how about its performance to directly segment CT images in target do- main? This setting, namely generalizable cross-modality segmentation, owning its clinical potential, is much more challenging than other related settings, e.g., domain adap- tation. To achieve this goal, we in this paper propose a novel dual-normalization module by leveraging the aug- mented source-similar and source-dissimilar images dur- ing our generalizable segmentation. To be specific, given a single source domain, aiming to simulate the possible appearance change in unseen target domains, we first uti- lize a nonlinear transformation to augment source-similar and source-dissimilar images. Then, to sufficiently ex- ploit these two types of augmentations, our proposed dual- normalization based model employs a shared backbone yet independent batch normalization layer for separate nor- malization. Afterwards, we put forward a style-based selec- tion scheme to automatically choose the appropriate path in the test stage. Extensive experiments on three publicly available datasets, i.e., BraTS, Cross-Modality Cardiac and Abdominal Multi-Organ dataset, have demonstrated that our method outperforms other state-of-the-art domain gen- eralization methods.
 ● 论文链接:https://arxiv.org/abs/2112.11177
● 论文链接:https://arxiv.org/abs/2112.11177
● 论文代码:https://github.com/zzzqzhou/Dual-Normalization
● 作者单位:南京大学,深圳大学,东南大学
[2] ACPL: Anti-curriculum Pseudo-labelling for Semi-supervised Medical Image Classification(半监督医学图像分类的反课程伪标签)
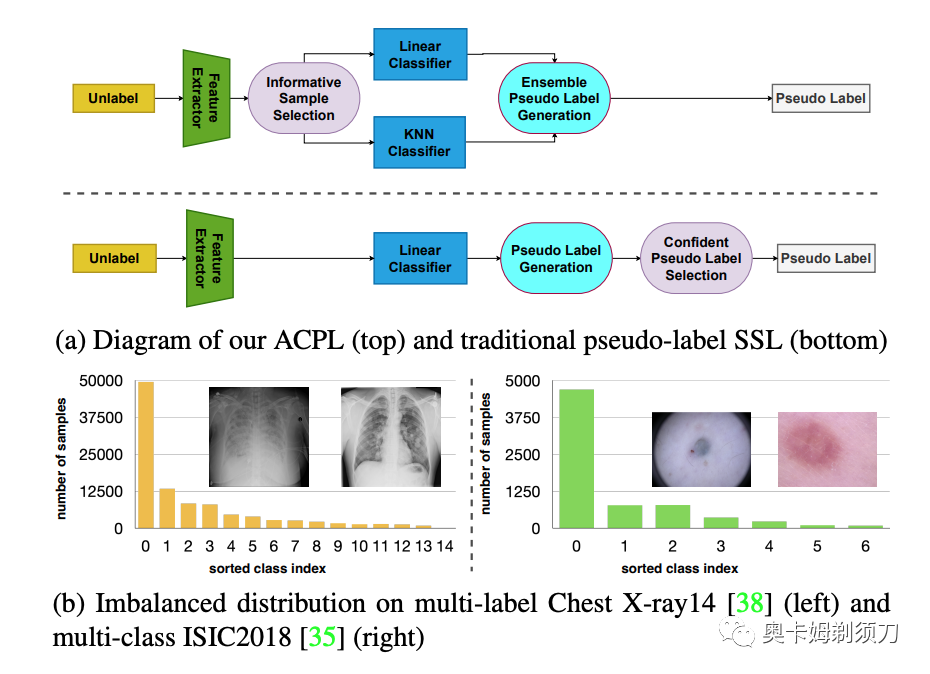
摘要:Effective semi-supervised learning (SSL) in medical image analysis (MIA) must address two challenges: 1) work effectively on both multi-class (e.g., lesion classification) and multi-label (e.g., multiple-disease diagnosis) problems, and 2) handle imbalanced learning (because of the high variance in disease prevalence). One strategy to explore in SSL MIA is based on the pseudo labelling strategy, but it has a few shortcomings. Pseudo-labelling has in general lower accuracy than consistency learning, it is not specifically designed for both multi-class and multi-label problems, and it can be challenged by imbalanced learning. In this paper, unlike traditional methods that select confident pseudo label by threshold, we propose a new SSL algorithm, called anti-curriculum pseudo-labelling (ACPL), which introduces novel techniques to select informative unlabelled samples, improving training balance and allowing the model to work for both multi-label and multi-class problems, and to estimate pseudo labels by an accurate ensemble of classifiers (improving pseudo label accuracy). We run extensive experiments to evaluate ACPL on two public medical image classification benchmarks: Chest X-Ray14 for thorax disease multi-label classification and ISIC2018 for skin lesion multi-class classification. Our method outperforms previous SOTA SSL methods on both datasets● 论文链接:https://arxiv.org/abs/2111.12918
● 作者单位:澳大利亚机器学习研究所,乌尔姆大学
[3] DTFD-MIL: Double-Tier Feature Distillation Multiple Instance Learning for Histopathology Whole Slide Image Classification(用于组织病理学全幻灯片图像分类的双层特征蒸馏多实例学习)
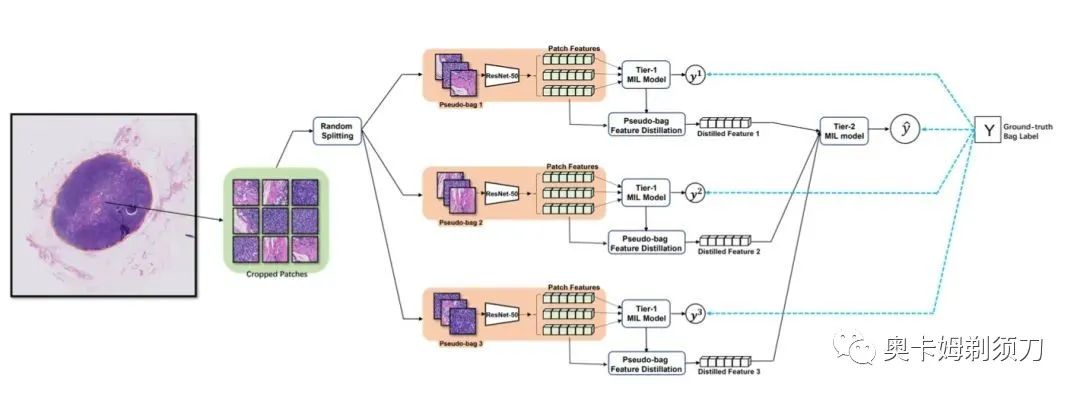
摘要:Multiple instance learning (MIL) has been increasingly used in the classification of histopathology whole slide images (WSIs). However, MIL approaches for this specific classification problem still face unique challenges, particularly those related to small sample cohorts. In these, there are limited number of WSI slides (bags), while the resolution of a single WSI is huge, which leads to a large number of patches (instances) cropped from this slide. To address this issue, we propose to virtually enlarge the number of bags by introducing the concept of pseudo-bags, on which a double-tier MIL framework is built to effectively use the intrinsic features. Besides, we also contribute to deriving the instance probability under the framework of attentionbased MIL, and utilize the derivation to help construct and analyze the proposed framework. The proposed method outperforms other latest methods on the CAMELYON-16 by substantially large margins, and is also better in performance on the TCGA lung cancer dataset. The proposed framework is ready to be extended for wider MIL applications.
● 论文链接:https://arxiv.org/abs/2203.12081
● 论文代码:https://github.com/hrzhang1123/DTFD-MIL
● 作者单位:利物浦大学,慈溪医工院等
[4] Robust Equivariant Imaging: a fully unsupervised framework for learning to image from noisy and partial measurements Transformer(鲁棒等变成像:一个学习从噪声和部分测量中成像的完全无监督框架)
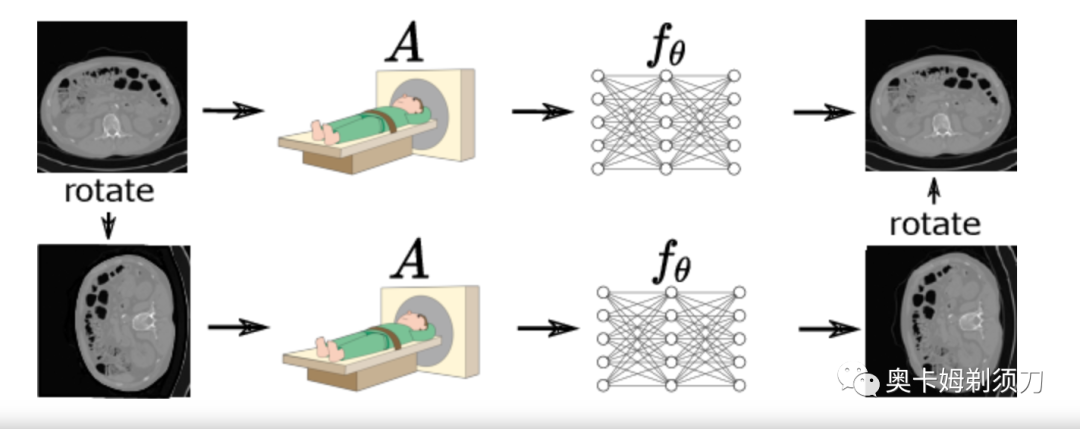
摘要:Deep networks provide state-of-the-art performance in multiple imaging inverse problems ranging from medical imaging to computational photography. However, most existing networks are trained with clean signals which are often hard or impossible to obtain. Equivariant imaging (EI) is a recent self-supervised learning framework that exploits the group invariance present in signal distributions to learn a reconstruction function from partial measurement data alone. While EI results are impressive, its performance degrades with increasing noise. In this paper, we propose a Robust Equivariant Imaging (REI) framework which can learn to image from noisy partial measurements alone. The proposed method uses Stein's Unbiased Risk Estimator (SURE) to obtain a fully unsupervised training loss that is robust to noise. We show that REI leads to considerable performance gains on linear and nonlinear inverse problems, thereby paving the way for robust unsupervised imaging with deep networks.
● 论文链接:https://arxiv.org/pdf/2111.12855.pdf
● 论文代码:https://github.com/edongdongchen/REI
● 作者单位:爱丁堡大学
[5] Incremental Cross-view Mutual Distillation for Self-supervised Medical CT Synthesis(用于自监督医学 CT 合成的增量交叉视图相互蒸馏)
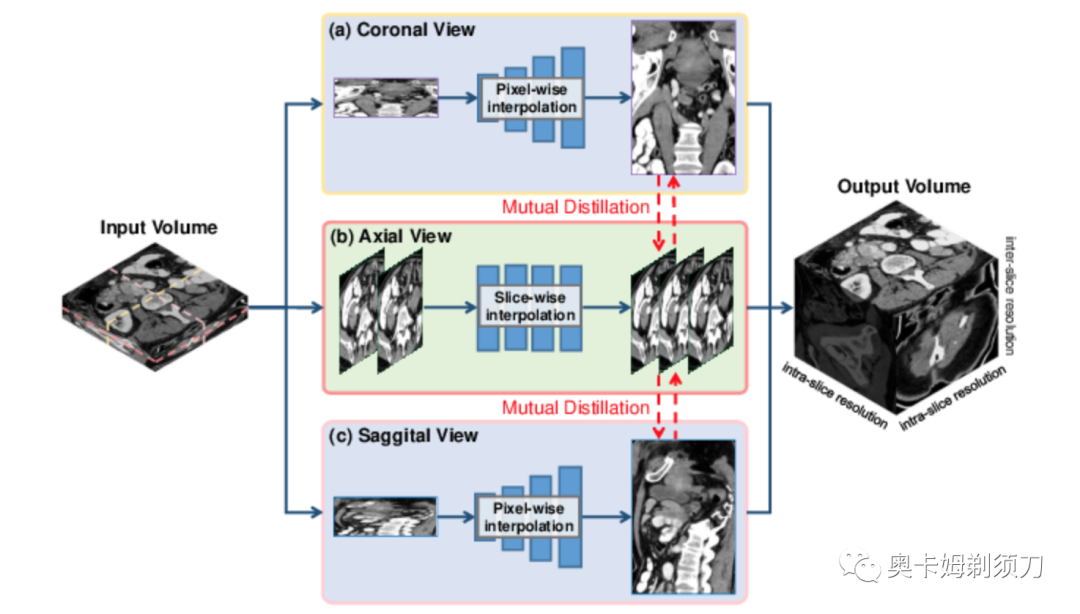
Due to the constraints of the imaging device and high cost in operation time, computer tomography (CT) scans are usually acquired with low intra-slice resolution. Improving the intra-slice resolution is beneficial to the disease diagnosis for both human experts and computer-aided systems. To this end, this paper builds a novel medical slice synthesis to increase the between-slice resolution. Considering that the ground-truth intermediate medical slices are always absent in clinical practice, we introduce the incremental cross-view mutual distillation strategy to accomplish this task in the self-supervised learning manner. Specifically, we model this problem from three different views: slice-wise interpolation from axial view and pixel-wise interpolation from coronal and sagittal views. Under this circumstance, the models learned from different views can distill valuable knowledge to guide the learning processes of each other. We can repeat this process to make the models synthesize intermediate slice data with increasing inter-slice resolution. To demonstrate the effectiveness of the proposed approach, we conduct comprehensive experiments on a large-scale CT dataset. Quantitative and qualitative comparison results show that our method outperforms state-of-the-art algorithms by clear margins.
● 论文链接:https://arxiv.org/abs/2112.10325
● 作者单位:西北工业大学
[6] ContIG: Self-supervised Multimodal Contrastive Learning for Medical Imaging with Genetics(基于遗传医学影像的自我监督多模态对比学习)
High annotation costs are a substantial bottleneck in applying modern deep learning architectures to clinically relevant medical use cases, substantiating the need for novel algorithms to learn from unlabeled data. In this work, we propose ContIG, a self-supervised method that can learn from large datasets of unlabeled medical images and genetic data. Our approach aligns images and several genetic modalities in the feature space using a contrastive loss. We design our method to integrate multiple modalities of each individual person in the same model end-to-end, even when the available modalities vary across individuals. Our procedure outperforms state-of-the-art self-supervised methods on all evaluated downstream benchmark tasks. We also adapt gradient-based explainability algorithms to better understand the learned cross-modal associations between the images and genetic modalities. Finally, we perform genome-wide association studies on the features learned by our models, uncovering interesting relationships between images and genetic data.
● 论文链接:https://arxiv.org/abs/2111.13424
● 作者单位:波茨坦大学等
[7] A variational Bayesian method for similarity learning in medical image registration(医学图像配准中相似度学习的变分贝叶斯方法)
● 作者:Daniel Grzech, Mohammad Farid Azampour, Ben Glocker, Julia Schnabel, Nassir Navab, Bernhard Kainz, Loic le Folgoc
● 作者单位:慕尼黑工业大学等
[8] Vox2Cortex: Fast Explicit Reconstruction of Cortical Surfaces from 3D MRI Scans with Geometric Deep Neural Networks(使用几何深度神经网络从 3D MRI 扫描中快速显式重建皮质表面)

The reconstruction of cortical surfaces from brain magnetic resonance imaging (MRI) scans is essential for quantitative analyses of cortical thickness and sulcal morphology. Although traditional and deep learning-based algorithmic pipelines exist for this purpose, they have two major drawbacks: lengthy runtimes of multiple hours (traditional) or intricate post-processing, such as mesh extraction and topology correction (deep learning-based). In this work, we address both of these issues and propose Vox2Cortex, a deep learning-based algorithm that directly yields topologically correct, three-dimensional meshes of the boundaries of the cortex. Vox2Cortex leverages convolutional and graph convolutional neural networks to deform an initial template to the densely folded geometry of the cortex represented by an input MRI scan. We show in extensive experiments on three brain MRI datasets that our meshes are as accurate as the ones reconstructed by state-of-the-art methods in the field, without the need for time- and resource-intensive post-processing. To accurately reconstruct the tightly folded cortex, we work with meshes containing about 168,000 vertices at test time, scaling deep explicit reconstruction methods to a new level.
● 论文链接:https://arxiv.org/abs/2203.09446
● 论文代码:https://github.com/ai-med/Vox2Cortex
● 作者单位:慕尼黑工业大学等
[9] Temporal Context Matters: Enhancing Single Image Prediction with Disease Progression Representations(时间上下文很重要:使用疾病进展表示增强单图像预测)
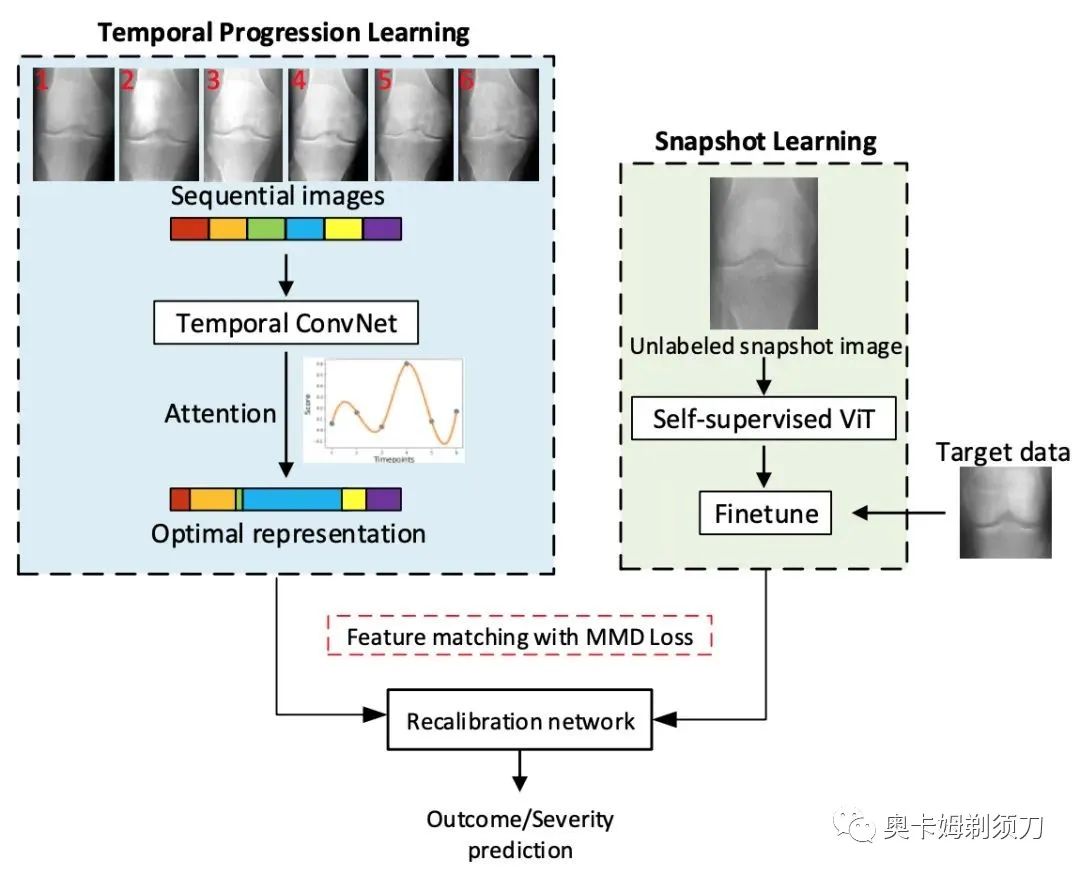
摘要:Clinical outcome or severity prediction from medical im- ages has largely focused on learning representations from single-timepoint or snapshot scans. It has been shown that disease progression can be better characterized by tempo- ral imaging. We therefore hypothesized that outcome pre- dictions can be improved by utilizing the disease progres- sion information from sequential images. We present a deep learning approach that leverages temporal progression in- formation to improve clinical outcome predictions from single-timepoint images. In our method, a self-attention based Temporal Convolutional Network (TCN) is used to learn a representation that is most reflective of the disease trajectory. Meanwhile, a Vision Transformer is pretrained in a self-supervised fashion to extract features from single- timepoint images. The key contribution is to design a recal- ibration module that employs maximum mean discrepancy loss (MMD) to align distributions of the above two contex- tual representations. We train our system to predict clini- cal outcomes and severity grades from single-timepoint im- ages. Experiments on chest and osteoarthritis radiography datasets demonstrate that our approach outperforms other state-of-the-art techniques.
● 论文链接:https://arxiv.org/abs/2203.01933
● 作者单位:石溪大学
[10] BoostMIS: Boosting Medical Image Semi-supervised Learning with Adaptive Pseudo Labeling and Informative Active Annotation( 医学图像半监督学习(SSL)框架)
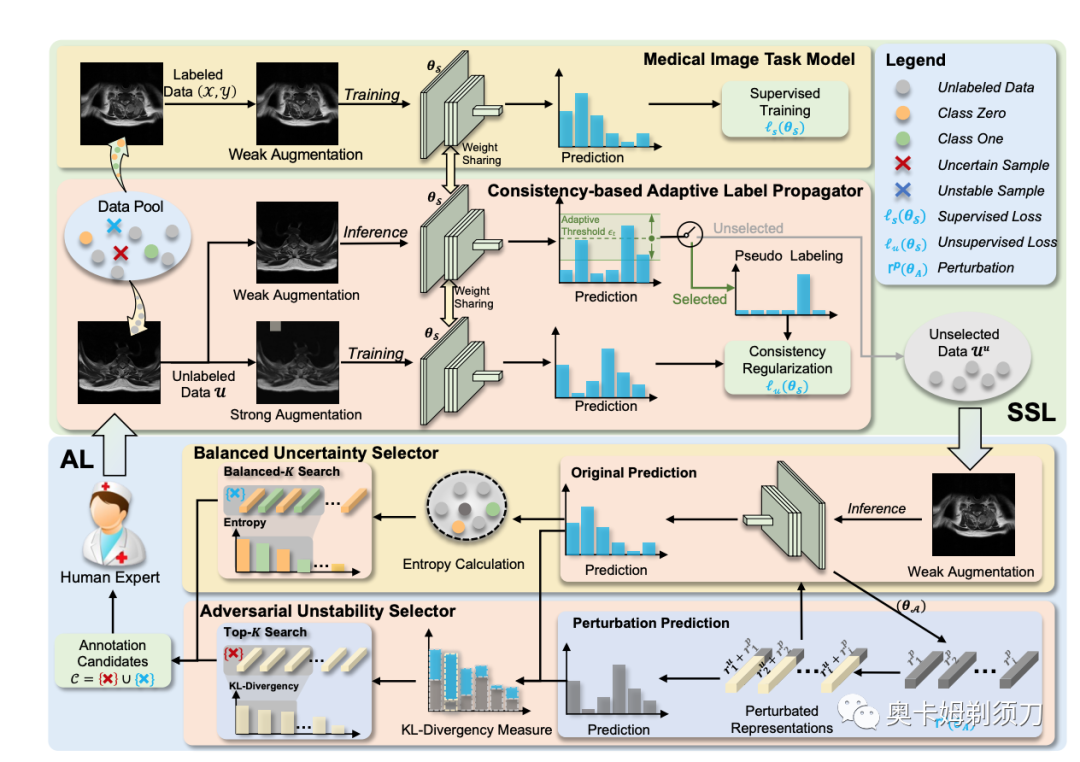
In this paper, we propose a novel semi-supervised learning (SSL) framework named BoostMIS that combines adaptive pseudo labeling and informative active annotation to unleash the potential of medical image SSL models: (1) BoostMIS can adaptively leverage the cluster assumption and consistency regularization of the unlabeled data according to the current learning status. This strategy can adaptively generate one-hot ``hard'' labels converted from task model predictions for better task model training. (2) For the unselected unlabeled images with low confidence, we introduce an Active learning (AL) algorithm to find the informative samples as the annotation candidates by exploiting virtual adversarial perturbation and model's density-aware entropy. These informative candidates are subsequently fed into the next training cycle for better SSL label propagation. Notably, the adaptive pseudo-labeling and informative active annotation form a learning closed-loop that are mutually collaborative to boost medical image SSL. To verify the effectiveness of the proposed method, we collected a metastatic epidural spinal cord compression (MESCC) dataset that aims to optimize MESCC diagnosis and classification for improved specialist referral and treatment. We conducted an extensive experimental study of BoostMIS on MESCC and another public dataset COVIDx. The experimental results verify our framework's effectiveness and generalisability for different medical image datasets with a significant improvement over various state-of-the-art methods.
● 论文链接:https://arxiv.org/abs/2203.02533
● 作者单位:NUS, 新加坡国立大学医院, 浙大
[11] Self-Supervised Pre-Training of Swin Transformers for 3D Medical Image Analysis(用于 3D 医学图像分析的 Swin Transformers 的自监督预训练)
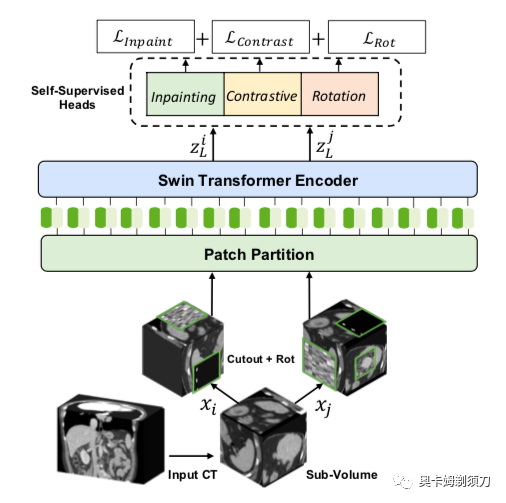
Vision Transformers (ViT)s have shown great performance in self-supervised learning of global and local representations that can be transferred to downstream applications. Inspired by these results, we introduce a novel self-supervised learning framework with tailored proxy tasks for medical image analy- sis. Specifically, we propose: (i) a new 3D transformer-based model, dubbed Swin UNEt TRansformers (Swin UNETR), with a hierarchical encoder for self-supervised pre-training; (ii) tailored proxy tasks for learning the underlying pattern of human anatomy. We demonstrate successful pre-training of the proposed model on 5,050 publicly available computed tomography (CT) images from various body organs. The ef- fectiveness of our approach is validated by fine-tuning the pre-trained models on the Beyond the Cranial Vault (BTCV) Segmentation Challenge with 13 abdominal organs and seg- mentation tasks from the Medical Segmentation Decathlon (MSD) dataset. Our model is currently the state-of-the-art on the public test leaderboards of both MSD and BTCV datasets. Code: https://monai.io/research/swin-unetr.
● 论文链接:https://arxiv.org/abs/2111.14791● 论文代码:https://monai.io/research/swin-unetr
● 作者单位:NVIDIA
[12] Adaptive Early-Learning Correction for Segmentation from Noisy Annotations(从噪声标签中分割的自适应早期学习校正)
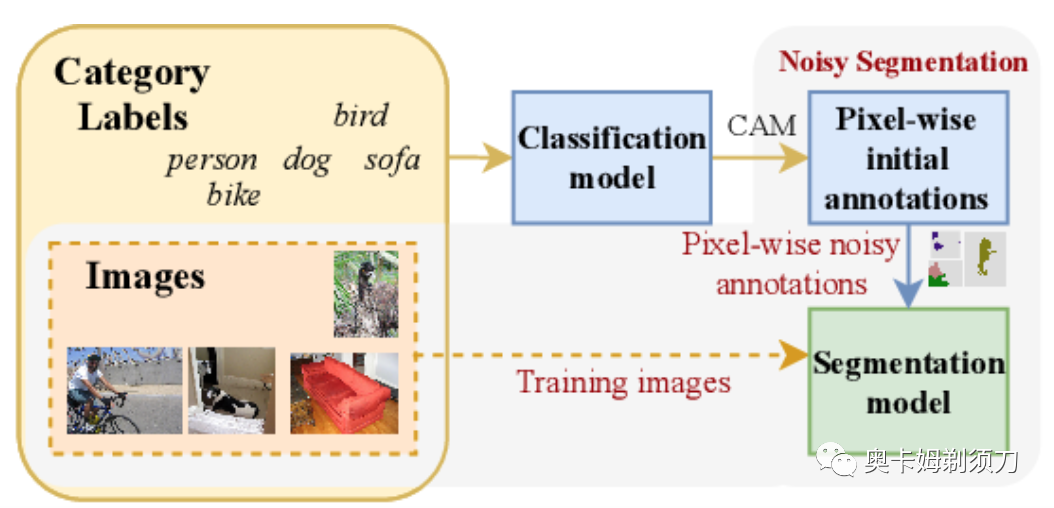
Deep learning in the presence of noisy annotations has been studied extensively in classification, but much less in segmentation tasks. In this work, we study the learning dynamics of deep segmentation networks trained on inaccurately-annotated data. We discover a phenomenon that has been previously reported in the context of classification: the networks tend to first fit the clean pixel-level labels during an "early-learning" phase, before eventually memorizing the false annotations. However, in contrast to classification, memorization in segmentation does not arise simultaneously for all semantic categories. Inspired by these findings, we propose a new method for segmentation from noisy annotations with two key elements. First, we detect the beginning of the memorization phase separately for each category during training. This allows us to adaptively correct the noisy annotations in order to exploit early learning. Second, we incorporate a regularization term that enforces consistency across scales to boost robustness against annotation noise. Our method outperforms standard approaches on a medical-imaging segmentation task where noises are synthesized to mimic human annotation errors. It also provides robustness to realistic noisy annotations present in weakly-supervised semantic segmentation, achieving state-of-the-art results on PASCAL VOC 2012
● 论文链接: https://arxiv.org/abs/2110.03740
● 论文代码: https://github.com/Kangningthu/ADELE
[13] Affine Medical Image Registration with Coarse-to-Fine Vision Transformer(基于粗-精视觉Transformer的仿射医学图像配准)
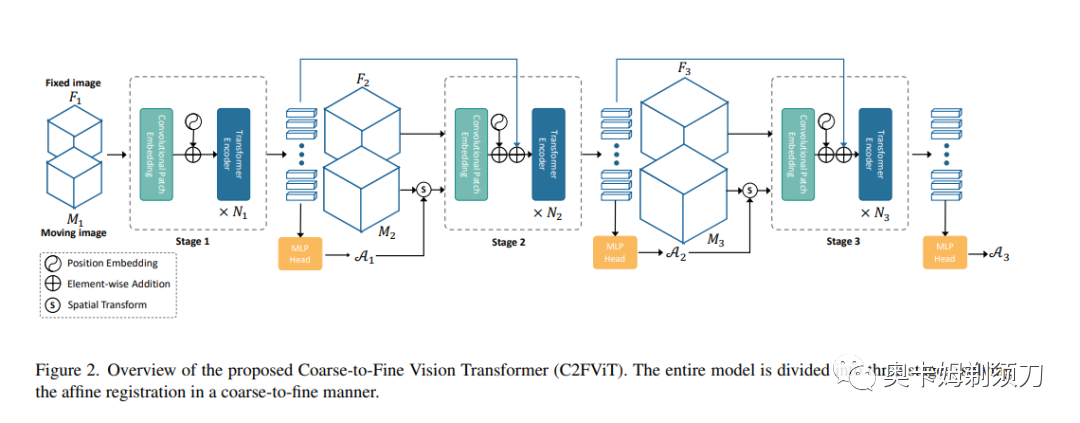
Affine registration is indispensable in a comprehensive medical image registration pipeline. However, only a few studies focus on fast and robust affine registration algorithms. Most of these studies utilize convolutional neural networks (CNNs) to learn joint affine and non-parametric registration, while the standalone performance of the affine subnetwork is less explored. Moreover, existing CNN-based affine registration approaches focus either on the local misalignment or the global orientation and position of the input to predict the affine transformation matrix, which are sensitive to spatial initialization and exhibit limited generalizability apart from the training dataset. In this paper, we present a fast and robust learning-based algorithm, Coarse-to-Fine Vision Transformer (C2FViT), for 3D affine medical image registration. Our method naturally leverages the global connectivity and locality of the convolutional vision transformer and the multi-resolution strategy to learn the global affine registration. We evaluate our method on 3D brain atlas registration and template-matching normalization. Comprehensive results demonstrate that our method is superior to the existing CNNs-based affine registration methods in terms of registration accuracy, robustness and generalizability while preserving the runtime advantage of the learning-based methods.
● 论文链接: https://arxiv.org/abs/2203.15216
● 论文代码: https://github.com/cwmok/C2FViT
● 作者单位:香港科技大学