Python也能画漂亮的complex heatmap?
微信公众号:「Computational Epigenetics」
关注生物信息学和计算表观遗传学。问题或建议,请公众号留言。
对于经常用R语言来画图的科研工作者来说,
应该对
ComplexHeatmap
(https://jokergoo.github.io/ComplexHeatmap-reference/book/)很
熟悉了吧。
这个包画的热图,既专业又漂亮。
可惜的是,在python中,一直没能出现一个可以画出好看complex heatmap的包,由于我们在用python做机器学习或者处理大数据的时候,也需要画热图,而在python和R中来回切换,也比较麻烦而且没有效率。
今天,给大家介绍一款可以在python中画出类似于R中ComplexHeatmap效果的包:
PyComplexHeatmap
(https://github.com/DingWB/PyComplexHeatmap)。
直接看下面的代码和图吧(教程来自:
https://github.com/DingWB/PyComplexHeatmap/blob/main/examples.ipynb):
1. 导入相关包
import os,sys
import PyComplexHeatmap
from PyComplexHeatmap import *
%matplotlib inline
import matplotlib.pylab as plt
plt.rcParams['figure.dpi'] = 120
plt.rcParams['savefig.dpi']=300
2. 快速入门
#Generate example dataset
df = pd.DataFrame(['AAAA1'] * 5 + ['BBBBB2'] * 5, columns=['AB'])
df['CD'] = ['C'] * 3 + ['D'] * 3 + ['G'] * 4
df['EF'] = ['E'] * 6 + ['F'] * 2 + ['H'] * 2
df['F'] = np.random.normal(0, 1, 10)
df.index = ['sample' + str(i) for i in range(1, df.shape[0] + 1)]
df_box = pd.DataFrame(np.random.randn(10, 4), columns=['Gene' + str(i) for i in range(1, 5)])
df_box.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_bar = pd.DataFrame(np.random.uniform(0, 10, (10, 2)), columns=['TMB1', 'TMB2'])
df_bar.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_scatter = pd.DataFrame(np.random.uniform(0, 10, 10), columns=['Scatter'])
df_scatter.index = ['sample' + str(i) for i in range(1, df_box.shape[0] + 1)]
df_heatmap = pd.DataFrame(np.random.randn(50, 10), columns=['sample' + str(i) for i in range(1, 11)])
df_heatmap.index = ["Fea" + str(i) for i in range(1, df_heatmap.shape[0] + 1)]
df_heatmap.iloc[1, 2] = np.nan
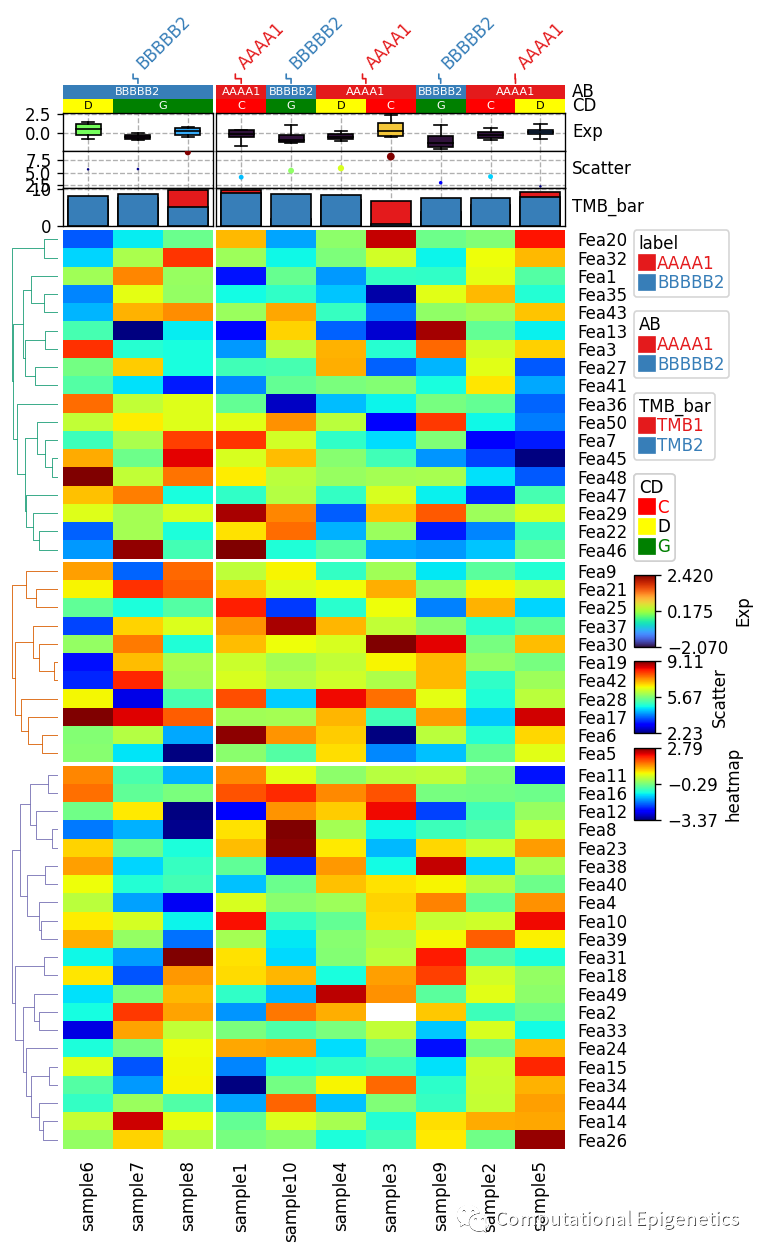
plt.figure(figsize=(6, 12))
row_ha = HeatmapAnnotation(label=anno_label(df.AB, merge=True),
AB=anno_simple(df.AB,add_text=True),axis=1,
CD=anno_simple(df.CD, colors={'C': 'red', 'D': 'yellow', 'G': 'green'},add_text=True),
Exp=anno_boxplot(df_box, cmap='turbo'),
Scatter=anno_scatterplot(df_scatter), TMB_bar=anno_barplot(df_bar),
)
cm = ClusterMapPlotter(data=df_heatmap, top_annotation=row_ha, col_split=2, row_split=3, col_split_gap=0.5,
row_split_gap=1,col_dendrogram=False,plot=True,
tree_kws={'col_cmap': 'Set1', 'row_cmap': 'Dark2'})
plt.savefig("example1_heatmap.pdf", bbox_inches='tight')
plt.show()
3. 画行/列注释
3.1 仅画行/列的注释信息
plt.figure(figsize=(6, 4))
row_ha = HeatmapAnnotation(label=anno_label(df.AB, merge=True),
AB=anno_simple(df.AB,add_text=True,legend=True), axis=1,
CD=anno_simple(df.CD, colors={'C': 'red', 'D': 'gray', 'G': 'yellow'},
add_text=True,legend=True),
Exp=anno_boxplot(df_box, cmap='turbo',legend=True),
Scatter=anno_scatterplot(df_scatter), TMB_bar=anno_barplot(df_bar,legend=True),
plot=True,legend=True,legend_gap=5
)
plt.savefig("col_annotation.pdf", bbox_inches='tight')
plt.show()
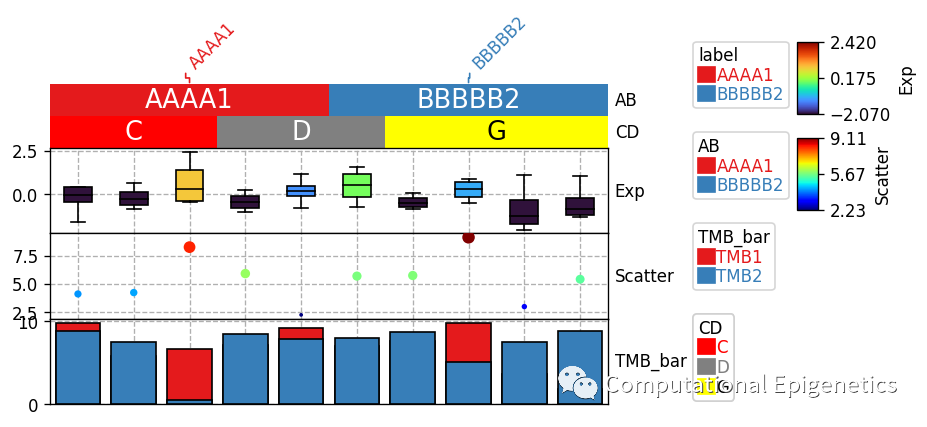
anno_label:
anno_label是用来将行/列注释信息(比如样本的性别、分组、亚型等)单独添加为一行文本(比如上图中倾斜的AAAA1和BBBBB2),merge参数控制是否将相邻两个或者多个单元格的注释信息合并为一个(如果相邻单元格的标签相同的话) 如果 merge != True, 那么,每一列的列标签都会被单独加上去(有时看起来会比较拥挤)。
anno_simple:
anno_simple是用来添加一个简单注释的函数(比如上图中的AB和CD那两列colorbar),cmap参数可以是分类型(categorical) (比如Set1, Dark2, tab10等) ,也可以是连续的 (比如jet, turbo, parula等)。 参数add_text 控制是否添加文本到单元格上面(比如上图中CD行单元格上面的文字C、D、G和AB列上面的注释文字)。如果颜色和字体大小没有被指定,函数会自动决定。比如,如果背景颜色是深色,那么文字颜色就会是浅色,否则字体颜色就是深色(比如CD行中的文字G就是被自动设定为黑色)。文字的颜色也可以通过参数text_kws={'color':your_color}来改变,比如:
plt.figure(figsize=(5, 4))
row_ha = HeatmapAnnotation(label=anno_label(df.AB, merge=True),
AB=anno_simple(df.AB,add_text=True,legend=True,text_kws={'color':'gold'}), axis=1,
CD=anno_simple(df.CD,add_text=True,legend=True,text_kws={'color':'purple'}),
Exp=anno_boxplot(df_box, cmap='turbo',legend=True),
Scatter=anno_scatterplot(df_scatter), TMB_bar=anno_barplot(df_bar,legend=True),
plot=True,legend=True,legend_gap=5)
plt.show()
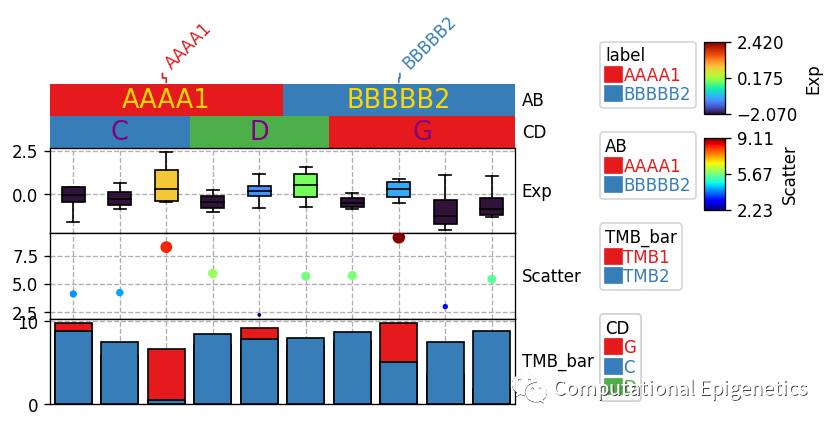
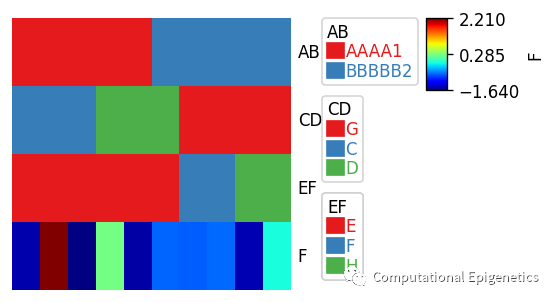
只需要一个python数据框dataframe就可以快速添加各类注释
当数据框df被给定时,该dataframe中的所有列都被单独作为anno_simple注释。比如,下面一个数据框df中有4列:AB、CD、EF、F,所有4列都会被自动画成列注释图。如果某一列不是连续型,而是字符等分类型变量,也可以用anno_boxplot或者anno_scatterplot等添加箱线图或者散点图作为列(比如样本)的信息注释(比如肿瘤样本的某种打分、某些基因表达的箱线图分布等)。
plt.figure(figsize=(3, 3))
row_ha = HeatmapAnnotation(df=df,plot=True,legend=True)
plt.show()
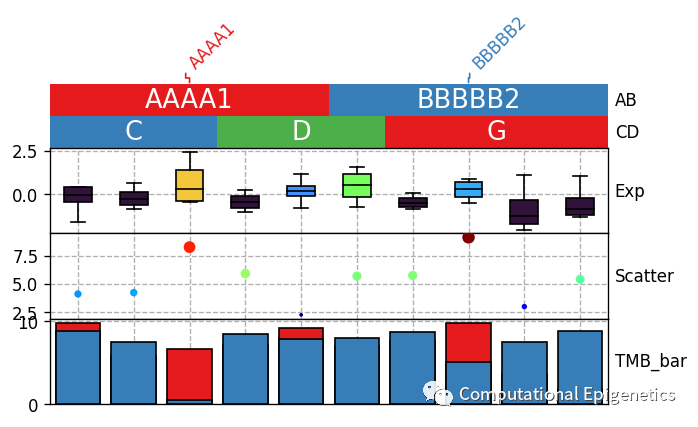
3.2 将图和图例分开
有时,我们可能会只需要图,不需要图例,也可能是要将图例单独画出来,PyComplexHeatmap可以实现这个功能,只需要让plot_legend=False,然后再新建一个图,执行 row_ha.plot_legends就可以单独画图例了。
只需要
plt.figure(figsize=(6, 4))
row_ha = HeatmapAnnotation(label=anno_label(df.AB, merge=True),
AB=anno_simple(df.AB,add_text=True,legend=True), axis=1,
CD=anno_simple(df.CD,add_text=True,legend=True),
Exp=anno_boxplot(df_box, cmap='turbo',legend=True),
Scatter=anno_scatterplot(df_scatter), TMB_bar=anno_barplot(df_bar,legend=True),
plot=True,legend=True,plot_legend=False,
legend_gap=5
)
plt.savefig("col_annotation.pdf", bbox_inches='tight')
plt.show()
plt.figure()
row_ha.plot_legends()
plt.savefig("legend.pdf",bbox_inches='tight')
plt.show()
No ax was provided, using plt.gca()

4. 画聚类图加行/列注释信息
我们这里使用 PyComplexHeatmap包中提供的example数据集:
!wget https://github.com/DingWB/PyComplexHeatmap/raw/main/data/influence_of_snp_on_beta.pickle
--2022-05-05 22:37:43-- https://github.com/DingWB/pyclustermap/raw/main/data/influence_of_snp_on_beta.pickle
Resolving github.com (github.com)... 140.82.112.4
Connecting to github.com (github.com)|140.82.112.4|:443... connected.
HTTP request sent, awaiting response... 404 Not Found
2022-05-05 22:37:43 ERROR 404: Not Found.
import pickle
import urllib
f=open("influence_of_snp_on_beta.pickle",'rb')
data=pickle.load(f)
f.close()
beta,snp,df_row,df_col,col_colors_dict,row_colors_dict=data
# beta is DNA methylation beta values matrix, df_row and df_col are row and columns annotation respectively, col_colors_dict and row_colors_dict are color for annotation
print(beta.iloc[:,list(range(5))].head(5))
print(df_row.head(5))
print(df_col.head(5))
beta=beta.sample(2000)
snp=snp.loc[beta.index.tolist()]
df_row=df_row.loc[beta.index.tolist()]
204875570030_R01C02 204875570030_R04C01 \
cg30848532_TC21 0.525089 0.419515
cg30147375_BC21 0.803776 0.585928
cg46239718_BC21 0.443958 0.517514
cg36100119_BC21 0.351977 0.528846
cg42738582_BC21 0.783958 0.724901
204875570030_R05C01 204875570030_R06C01 204875570035_R05C02
cg30848532_TC21 0.483276 0.460750 0.390317
cg30147375_BC21 0.510269 0.831463 0.550146
cg46239718_BC21 0.535909 0.450167 0.564107
cg36100119_BC21 0.524896 0.374422 0.551200
cg42738582_BC21 0.802178 0.848621 0.850481
chr Target CpG ExtensionBase ProbeDesign CON mapFlag \
cg30848532_TC21 chr12 1 1 0 II C 16
cg30147375_BC21 chr11 0 0 0 II C 0
cg46239718_BC21 chr8 1 1 0 II C 0
cg36100119_BC21 chr19 1 1 0 II C 16
cg42738582_BC21 chr5 0 0 0 II C 16
Group \
cg30848532_TC21 Suboptimal hybridization
cg30147375_BC21 No Effect
cg46239718_BC21 Artificial low meth. reading
cg36100119_BC21 Suboptimal hybridization
cg42738582_BC21 Suboptimal hybridization
Type
cg30848532_TC21 1-1-0-CG-GG-II-C-16-GA-chr12-79760438
cg30147375_BC21 0-0-0-ca-ac-II-C-0-AG-chr11-109557651
cg46239718_BC21 1-1-0-cg-gt-II-C-0-GA-chr8-117860829
cg36100119_BC21 1-1-0-CG-GG-II-C-16-GA-chr19-5877949
cg42738582_BC21 0-0-0-AA-AA-II-C-16-AG-chr5-122031379
Strain Tissue Sex
204875570030_R01C02 MOLF_EiJ Frontal Lobe Brain Female
204875570030_R04C01 CAST_EiJ Frontal Lobe Brain Male
204875570030_R05C01 CAST_EiJ Frontal Lobe Brain Female
204875570030_R06C01 MOLF_EiJ Frontal Lobe Brain Male
204875570035_R05C02 CAST_EiJ Liver Male
row_ha = HeatmapAnnotation(Target=anno_simple(df_row.Target,colors=row_colors_dict['Target'],rasterized=True),
Group=anno_simple(df_row.Group,colors=row_colors_dict['Group'],rasterized=True),
axis=0)
col_ha= HeatmapAnnotation(label=anno_label(df_col.Strain,merge=True,rotation=15),
Strain=anno_simple(df_col.Strain,add_text=True),
Tissue=df_col.Tissue,Sex=df_col.Sex,axis=1) #df=df_col.loc[:,['Strain','Tissue','Sex']]
plt.figure(figsize=(6, 10))
cm = ClusterMapPlotter(data=beta, top_annotation=col_ha, left_annotation=row_ha,
show_rownames=False,show_colnames=False,
row_dendrogram=False,col_dendrogram=False,
row_split=df_row.loc[:, ['Target', 'Group']],
col_split=df_col['Strain'],cmap='parula',
rasterized=True,row_split_gap=1,legend=True,
tree_kws={'col_cmap':'Set1'})
plt.savefig("clustermap.pdf", bbox_inches='tight')
plt.show()
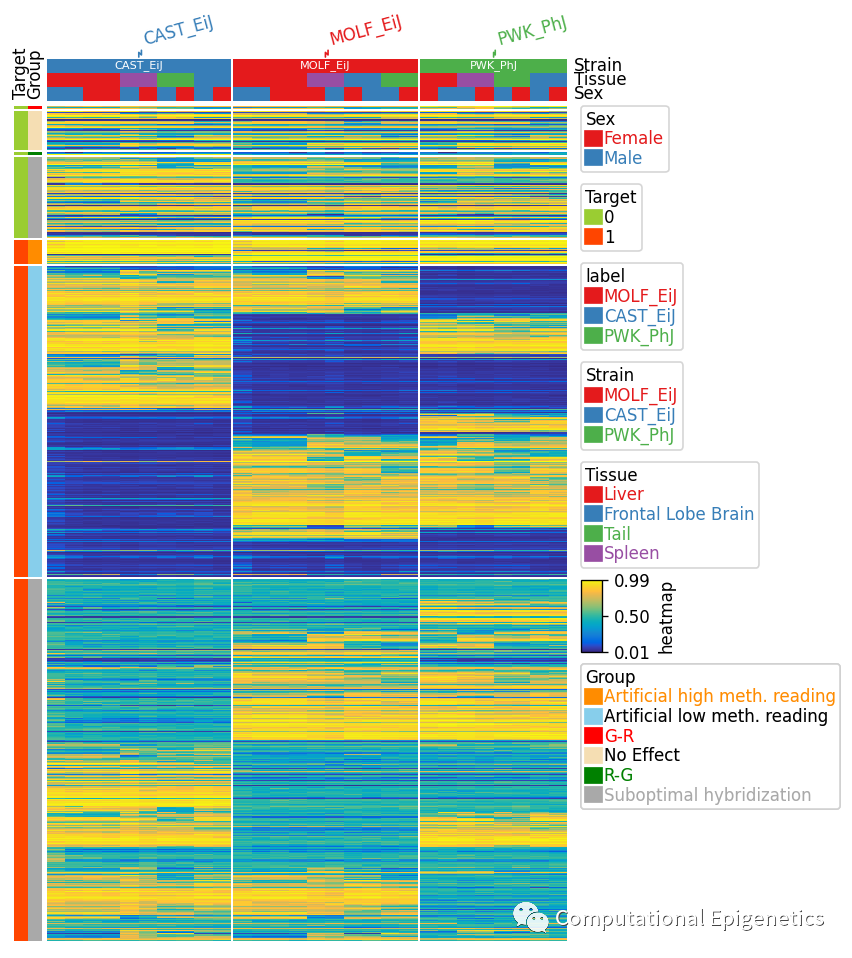
Key features:
用户可以通过row_split和col_split将所有的行和列按照标签分割成不同的模块,row_split and col_split 可以是数字(分成几个subgroup)、pandas dataframe或者是Series (每个样本对应的类别信息)。
5. 将多个热图[聚类图]水平或者垂直拼接起来
row_ha = HeatmapAnnotation(Target=anno_simple(df_row.Target, colors=row_colors_dict['Target'], rasterized=True),
Group=anno_simple(df_row.Group, colors=row_colors_dict['Group'], rasterized=True),
axis=0)
col_ha = HeatmapAnnotation(label=anno_label(df_col.Strain, merge=True, rotation=15),
Strain=anno_simple(df_col.Strain, add_text=True),
Tissue=df_col.Tissue, Sex=df_col.Sex,
axis=1) # df=df_col.loc[:,['Strain','Tissue','Sex']]
cm1 = ClusterMapPlotter(data=beta, top_annotation=col_ha, left_annotation=row_ha,
show_rownames=False, show_colnames=False,
row_dendrogram=False, col_dendrogram=False,
row_split=df_row.loc[:, ['Target', 'Group']],
col_split=df_col['Strain'], cmap='parula',
rasterized=True, row_split_gap=1, legend=True,
plot=False,label='beta',
tree_kws={'col_cmap': 'Set1'}) #
cm2 = ClusterMapPlotter(data=snp, top_annotation=col_ha, left_annotation=row_ha,
show_rownames=False, show_colnames=False,
row_dendrogram=False, col_dendrogram=False,
col_cluster_method='ward',row_cluster_method='ward',
col_cluster_metric='jaccard',row_cluster_metric='jaccard',
row_split=df_row.loc[:, ['Target', 'Group']],
col_split=df_col['Strain'],
rasterized=True, row_split_gap=1, legend=True,
plot=False,cmap='Greys',label='SNP',
tree_kws={'col_cmap': 'Set1'}) #
cmlist=[cm1,cm2]
plt.figure(figsize=(10,12))
composite(cmlist=cmlist, main=1,legendpad=0,legend_y=0.8)
plt.savefig("beta_snp.pdf", bbox_inches='tight')
plt.show()

希望这篇文章能对大家有帮助!扫描文末二维码或者搜索关注 Computational Epigenetics 公众号,我们会经常分享生物信息学和计算表观遗传学相关的文章。
往期精品(点击图片直达文字对应教程)
后台回复“
生信宝典福利第一波
”或点击
阅读原文
获取教程合集