【干货】特征选择的通俗讲解!
收集的数据格式不对(如 SQL 数据库、JSON、CSV 等)
缺失值和异常值
标准化
减少数据集中存在的固有噪声(部分存储数据可能已损坏)
数据集中的某些功能可能无法收集任何信息以供分析
提高精度
降低过拟合风险
加快训练速度
改进数据可视化
增加我们模型的可解释性

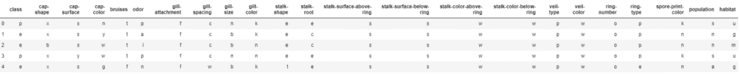
X = df.drop(['class'], axis = 1)Y = df['class']X = pd.get_dummies(X, prefix_sep='_')Y = LabelEncoder().fit_transform(Y)X2 = StandardScaler().fit_transform(X)X_Train, X_Test, Y_Train, Y_Test = train_test_split(X2, Y, test_size = 0.30, random_state = 101)
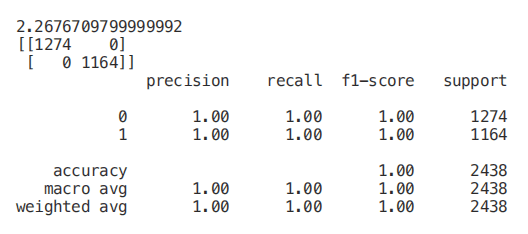
start = time.process_time()trainedforest = RandomForestClassifier(n_estimators=700).fit(X_Train,Y_Train)print(time.process_time() - start)predictionforest = trainedforest.predict(X_Test)print(confusion_matrix(Y_Test,predictionforest))print(classification_report(Y_Test,predictionforest))
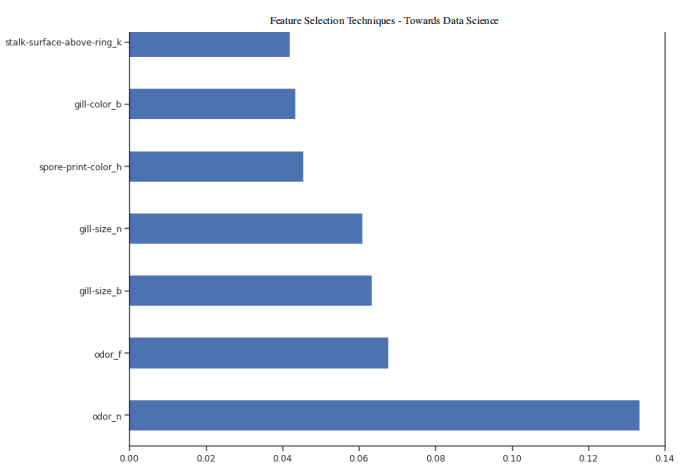
figure(num=None, figsize=(20, 22), dpi=80, facecolor='w', edgecolor='k')feat_importances = pd.Series(trainedforest.feature_importances_, index= X.columns)feat_importances.nlargest(7).plot(kind='barh')
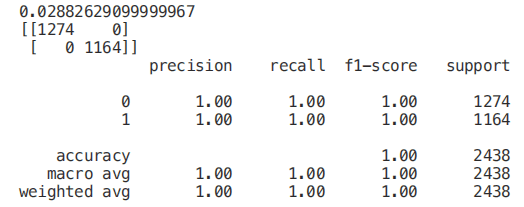
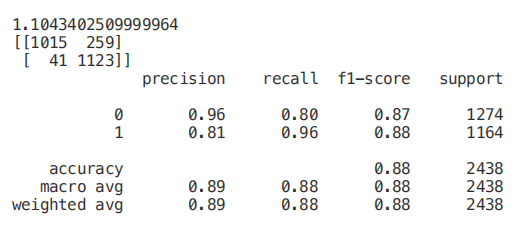
X_Reduced = X[['odor_n','odor_f', 'gill-size_n','gill-size_b']]X_Reduced = StandardScaler().fit_transform(X_Reduced)X_Train2, X_Test2, Y_Train2, Y_Test2 = train_test_split(X_Reduced, Y, test_size = 0.30, random_state = 101)start = time.process_time()trainedforest = RandomForestClassifier(n_estimators=700).fit(X_Train2,Y_Train2)print(time.process_time() - start)predictionforest = trainedforest.predict(X_Test2)print(confusion_matrix(Y_Test2,predictionforest))print(classification_report(Y_Test2,predictionforest))
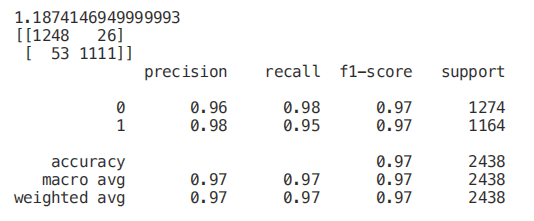
start = time.process_time()trainedtree = tree.DecisionTreeClassifier().fit(X_Train, Y_Train)print(time.process_time() - start)predictionstree = trainedtree.predict(X_Test)print(confusion_matrix(Y_Test,predictionstree))print(classification_report(Y_Test,predictionstree))
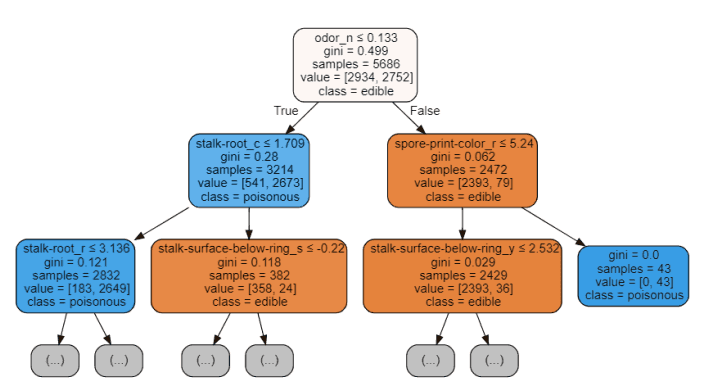
import graphvizfrom sklearn.tree import DecisionTreeClassifier, export_graphvizdata = export_graphviz(trainedtree,out_file=None,feature_names= X.columns,class_names=['edible', 'poisonous'],filled=True, rounded=True,max_depth=2,special_characters=True)graph = graphviz.Source(data)graph
from sklearn.feature_selection import RFEmodel = RandomForestClassifier(n_estimators=700)rfe = RFE(model, 4)start = time.process_time()RFE_X_Train = rfe.fit_transform(X_Train,Y_Train)RFE_X_Test = rfe.transform(X_Test)rfe = rfe.fit(RFE_X_Train,Y_Train)print(time.process_time() - start)print("Overall Accuracy using RFE: ", rfe.score(RFE_X_Test,Y_Test))
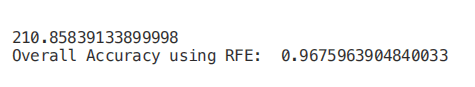
SelecFromModel
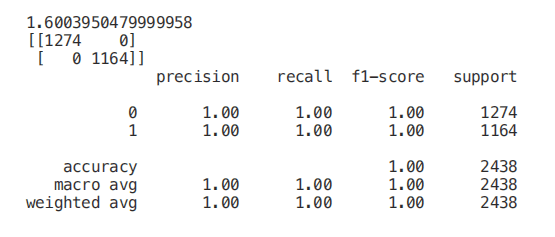
from sklearn.ensemble import ExtraTreesClassifierfrom sklearn.feature_selection import SelectFromModelmodel = ExtraTreesClassifier()start = time.process_time()model = model.fit(X_Train,Y_Train)model = SelectFromModel(model, prefit=True)print(time.process_time() - start)Selected_X = model.transform(X_Train)start = time.process_time()trainedforest = RandomForestClassifier(n_estimators=700).fit(Selected_X, Y_Train)print(time.process_time() - start)Selected_X_Test = model.transform(X_Test)predictionforest = trainedforest.predict(Selected_X_Test)print(confusion_matrix(Y_Test,predictionforest))print(classification_report(Y_Test,predictionforest))
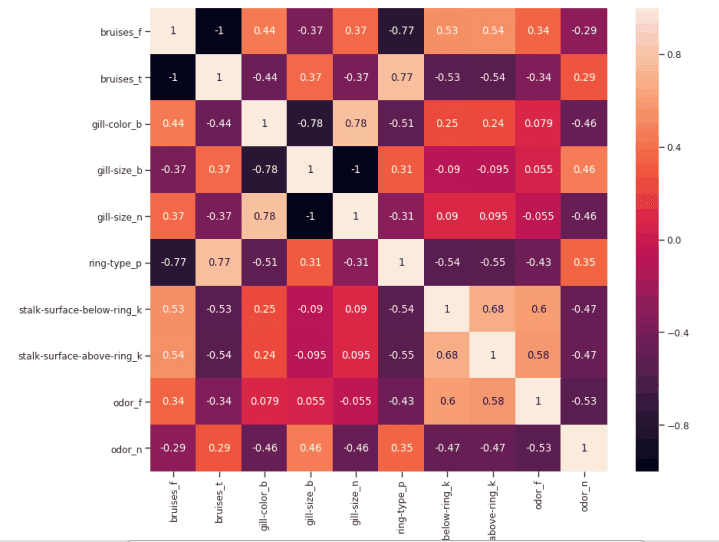
如果两个特征之间的相关性为 0,则意味着更改这两个特征中的任何一个都不会影响另一个。
如果两个特征之间的相关性大于 0,这意味着增加一个特征中的值也会增加另一个特征中的值(相关系数越接近 1,两个不同特征之间的这种联系就越强)。
如果两个特征之间的相关性小于 0,这意味着增加一个特征中的值将使减少另一个特征中的值(相关性系数越接近-1,两个不同特征之间的这种关系将越强)。
Numeric_df = pd.DataFrame(X)Numeric_df['Y'] = Ycorr= Numeric_df.corr()corr_y = abs(corr["Y"])highest_corr = corr_y[corr_y >0.5]highest_corr.sort_values(ascending=True)
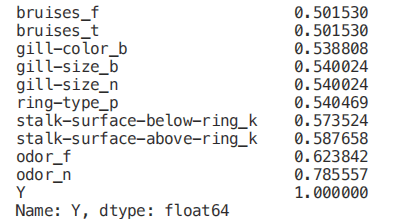
figure(num=None, figsize=(12, 10), dpi=80, facecolor='w', edgecolor='k')corr2 = Numeric_df[['bruises_f' , 'bruises_t' , 'gill-color_b' , 'gill-size_b' , 'gill-size_n' , 'ring-type_p' , 'stalk-surface-below-ring_k' , 'stalk-surface-above-ring_k' , 'odor_f', 'odor_n']].corr()sns.heatmap(corr2, annot=True, fmt=".2g")
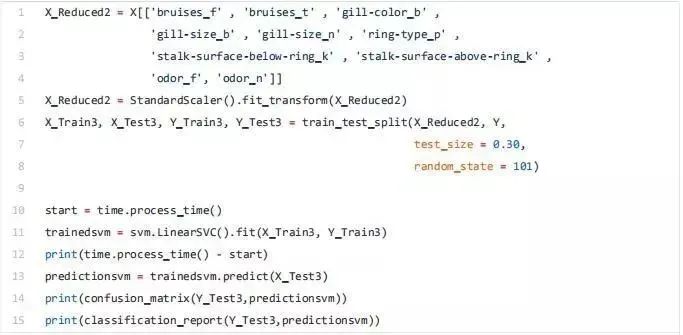
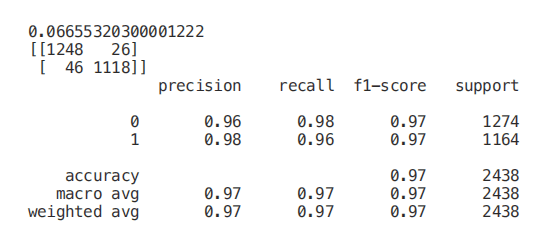
单变量选择
Classification = chi2, f_classif, mutual_info_classif
Regression = f_regression, mutual_info_regression
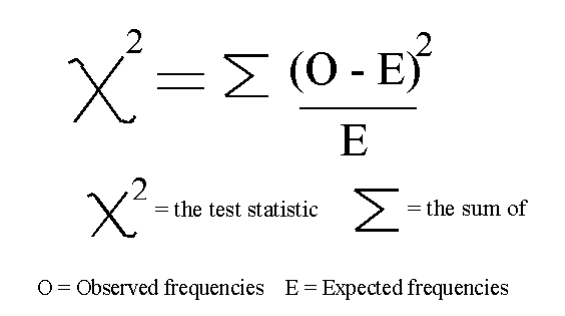
from sklearn.feature_selection import SelectKBestfrom sklearn.feature_selection import chi2min_max_scaler = preprocessing.MinMaxScaler()Scaled_X = min_max_scaler.fit_transform(X2)X_new = SelectKBest(chi2, k=2).fit_transform(Scaled_X, Y)X_Train3, X_Test3, Y_Train3, Y_Test3 = train_test_split(X_new, Y, test_size = 0.30, random_state = 101)start = time.process_time()trainedforest = RandomForestClassifier(n_estimators=700).fit(X_Train3,Y_Train3)print(time.process_time() - start)predictionforest = trainedforest.predict(X_Test3)print(confusion_matrix(Y_Test3,predictionforest))print(classification_report(Y_Test3,predictionforest))
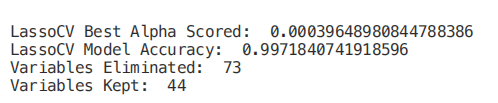
from sklearn.linear_model import LassoCVregr = LassoCV(cv=5, random_state=101)regr.fit(X_Train,Y_Train)print("LassoCV Best Alpha Scored: ", regr.alpha_)print("LassoCV Model Accuracy: ", regr.score(X_Test, Y_Test))model_coef = pd.Series(regr.coef_, index = list(X.columns[:-1]))print("Variables Eliminated: ", str(sum(model_coef == 0)))print("Variables Kept: ", str(sum(model_coef != 0)))
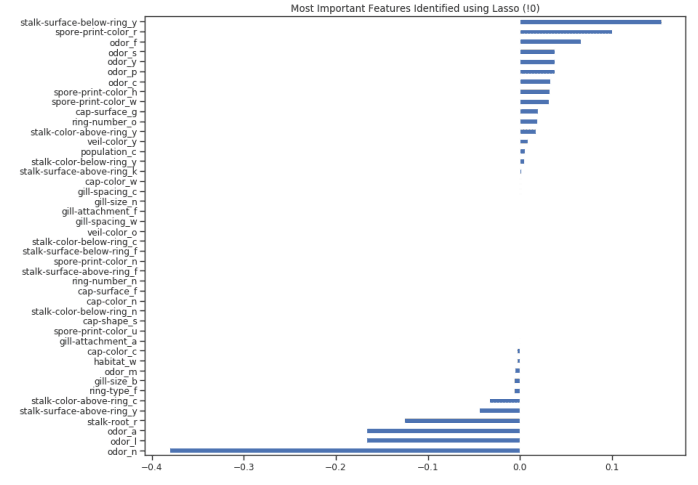
figure(num=None, figsize=(12, 10), dpi=80, facecolor='w', edgecolor='k')top_coef = model_coef.sort_values()top_coef[top_coef != 0].plot(kind = "barh")plt.title("Most Important Features Identified using Lasso (!0)")
评论