Bokeh,一个超强交互式Python可视化库!
今天这篇推文,给大家介绍一下Python中常用且可灵活交互使用的的可视化绘制包- Bokeh,由于网上关于该包较多及官方介绍也较为详细,这里就在不再过多介绍,我们直接放出几副精美的可视化作品供大家欣赏:
在jupyter notebook 中显示
在绘制可视化作品之前需输入:
output_notebook()
即可在jupyter notebook 中交互显示可视化结果。
Bokeh 可视化作品欣赏
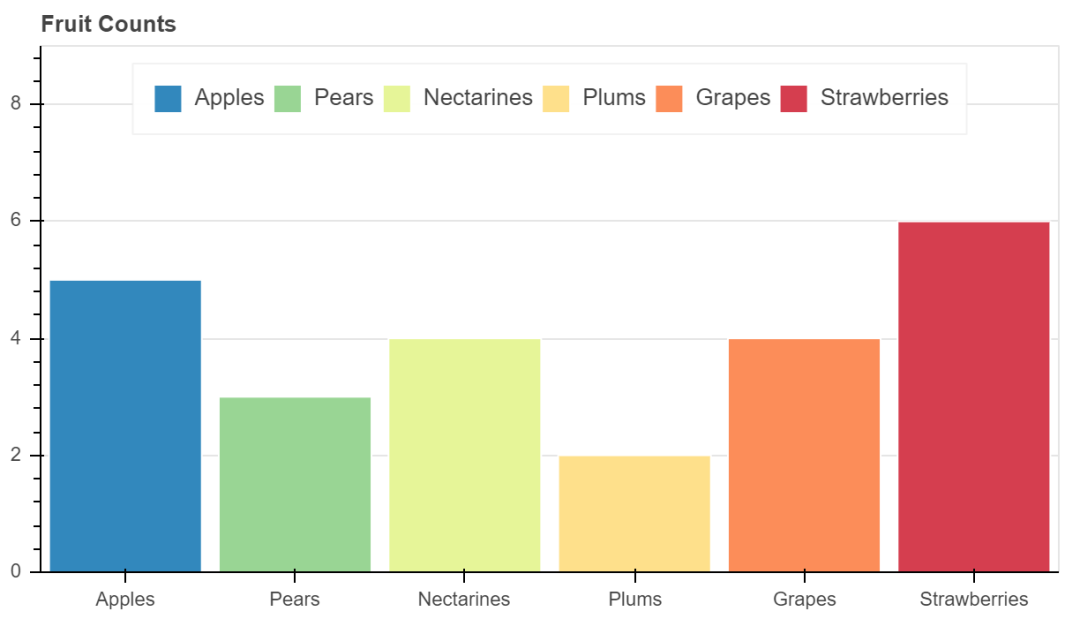
bar_colormapped
from bokeh.io import output_file, show
from bokeh.models import ColumnDataSource
from bokeh.palettes import Spectral6
from bokeh.plotting import figure
from bokeh.transform import factor_cmap
output_file("bar_colormapped.html")
fruits = ['Apples', 'Pears', 'Nectarines', 'Plums', 'Grapes', 'Strawberries']
counts = [5, 3, 4, 2, 4, 6]
source = ColumnDataSource(data=dict(fruits=fruits, counts=counts))
p = figure(x_range=fruits, plot_height=350, toolbar_location=None, title="Fruit Counts")
p.vbar(x='fruits', top='counts', width=0.9, source=source, legend_field="fruits",
line_color='white', fill_color=factor_cmap('fruits', palette=Spectral6, factors=fruits))
p.xgrid.grid_line_color = None
p.y_range.start = 0
p.y_range.end = 9
p.legend.orientation = "horizontal"
p.legend.location = "top_center"
show(p)
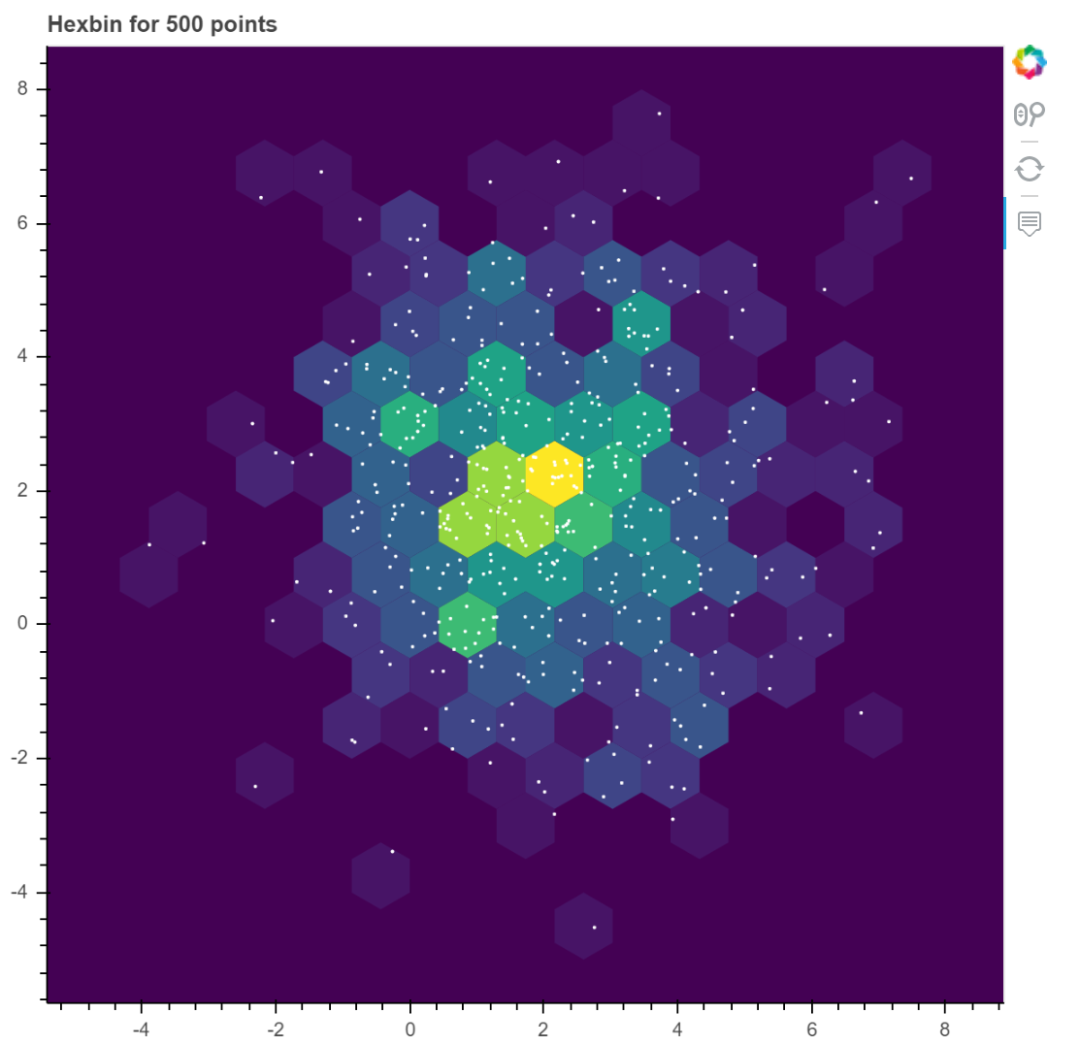
hexbin
import numpy as np
from bokeh.io import output_file, show
from bokeh.models import HoverTool
from bokeh.plotting import figure
n = 500
x = 2 + 2*np.random.standard_normal(n)
y = 2 + 2*np.random.standard_normal(n)
p = figure(title="Hexbin for 500 points", match_aspect=True,
tools="wheel_zoom,reset", background_fill_color='#440154')
p.grid.visible = False
r, bins = p.hexbin(x, y, size=0.5, hover_color="pink", hover_alpha=0.8)
p.circle(x, y, color="white", size=1)
p.add_tools(HoverTool(
tooltips=[("count", "@c"), ("(q,r)", "(@q, @r)")],
mode="mouse", point_policy="follow_mouse", renderers=[r]
))
output_file("hexbin.html")
show(p)
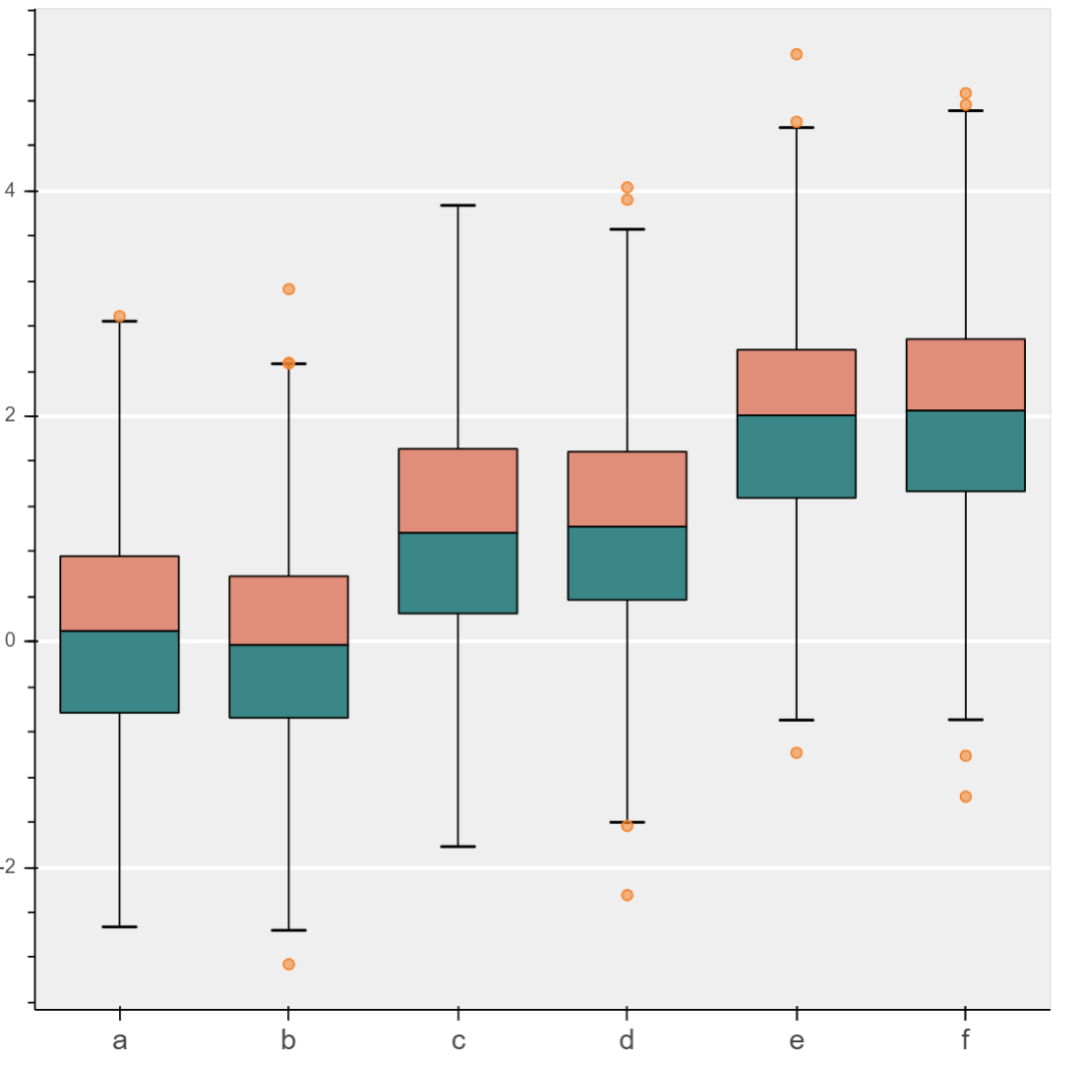
boxplot
import numpy as np
import pandas as pd
from bokeh.plotting import figure, output_file, show
# generate some synthetic time series for six different categories
cats = list("abcdef")
yy = np.random.randn(2000)
g = np.random.choice(cats, 2000)
for i, l in enumerate(cats):
yy[g == l] += i // 2
df = pd.DataFrame(dict(score=yy, group=g))
# find the quartiles and IQR for each category
groups = df.groupby('group')
q1 = groups.quantile(q=0.25)
q2 = groups.quantile(q=0.5)
q3 = groups.quantile(q=0.75)
iqr = q3 - q1
upper = q3 + 1.5*iqr
lower = q1 - 1.5*iqr
# find the outliers for each category
def outliers(group):
cat = group.name
return group[(group.score > upper.loc[cat]['score']) | (group.score < lower.loc[cat]['score'])]['score']
out = groups.apply(outliers).dropna()
# prepare outlier data for plotting, we need coordinates for every outlier.
if not out.empty:
outx = []
outy = []
for keys in out.index:
outx.append(keys[0])
outy.append(out.loc[keys[0]].loc[keys[1]])
p = figure(tools="", background_fill_color="#efefef", x_range=cats, toolbar_location=None)
# if no outliers, shrink lengths of stems to be no longer than the minimums or maximums
qmin = groups.quantile(q=0.00)
qmax = groups.quantile(q=1.00)
upper.score = [min([x,y]) for (x,y) in zip(list(qmax.loc[:,'score']),upper.score)]
lower.score = [max([x,y]) for (x,y) in zip(list(qmin.loc[:,'score']),lower.score)]
# stems
p.segment(cats, upper.score, cats, q3.score, line_color="black")
p.segment(cats, lower.score, cats, q1.score, line_color="black")
# boxes
p.vbar(cats, 0.7, q2.score, q3.score, fill_color="#E08E79", line_color="black")
p.vbar(cats, 0.7, q1.score, q2.score, fill_color="#3B8686", line_color="black")
# whiskers (almost-0 height rects simpler than segments)
p.rect(cats, lower.score, 0.2, 0.01, line_color="black")
p.rect(cats, upper.score, 0.2, 0.01, line_color="black")
# outliers
if not out.empty:
p.circle(outx, outy, size=6, color="#F38630", fill_alpha=0.6)
p.xgrid.grid_line_color = None
p.ygrid.grid_line_color = "white"
p.grid.grid_line_width = 2
p.xaxis.major_label_text_font_size="16px"
output_file("boxplot.html", title="boxplot.py example")
show(p)
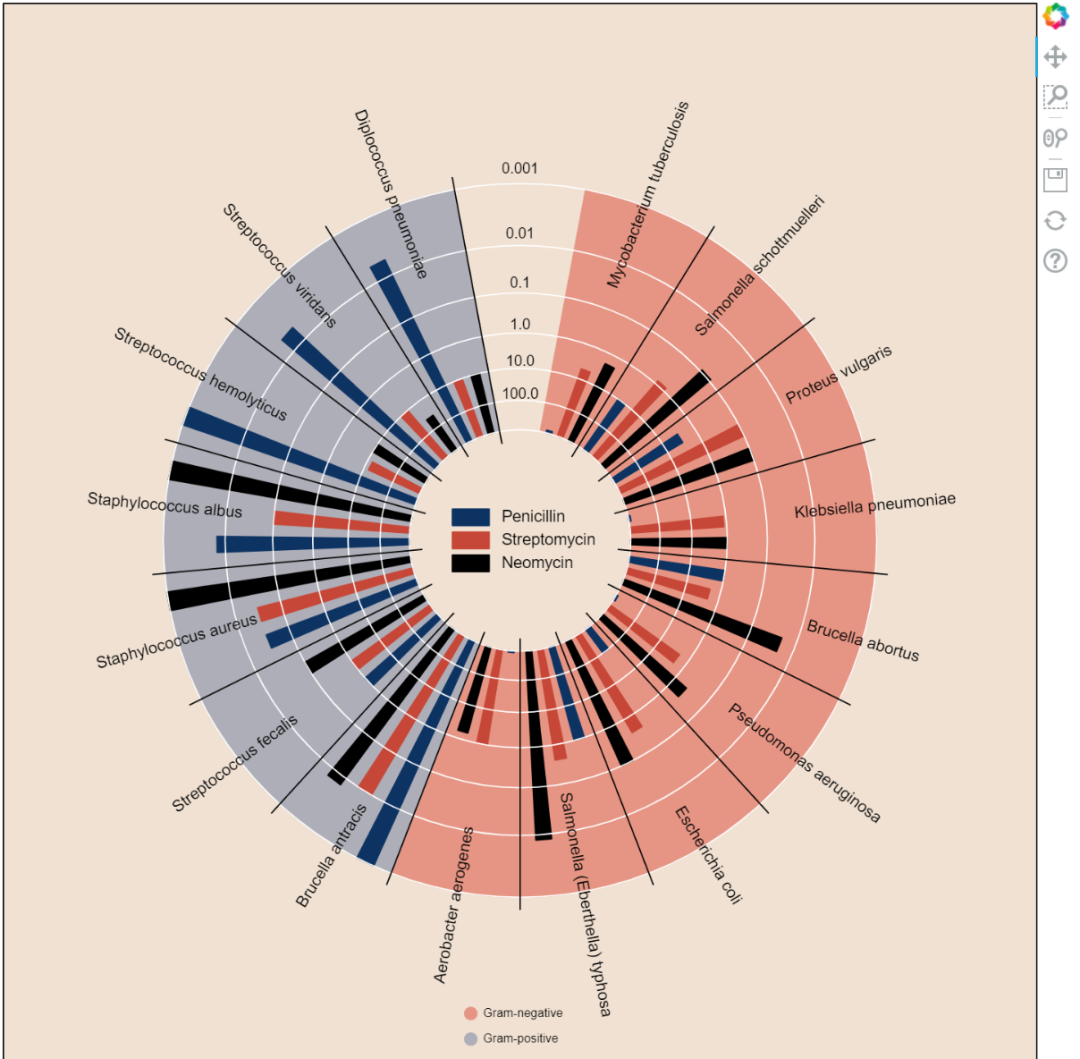
burtin
from collections import OrderedDict
from io import StringIO
from math import log, sqrt
import numpy as np
import pandas as pd
from bokeh.plotting import figure, output_file, show
antibiotics = """
bacteria, penicillin, streptomycin, neomycin, gram
Mycobacterium tuberculosis, 800, 5, 2, negative
Salmonella schottmuelleri, 10, 0.8, 0.09, negative
Proteus vulgaris, 3, 0.1, 0.1, negative
Klebsiella pneumoniae, 850, 1.2, 1, negative
Brucella abortus, 1, 2, 0.02, negative
Pseudomonas aeruginosa, 850, 2, 0.4, negative
Escherichia coli, 100, 0.4, 0.1, negative
Salmonella (Eberthella) typhosa, 1, 0.4, 0.008, negative
Aerobacter aerogenes, 870, 1, 1.6, negative
Brucella antracis, 0.001, 0.01, 0.007, positive
Streptococcus fecalis, 1, 1, 0.1, positive
Staphylococcus aureus, 0.03, 0.03, 0.001, positive
Staphylococcus albus, 0.007, 0.1, 0.001, positive
Streptococcus hemolyticus, 0.001, 14, 10, positive
Streptococcus viridans, 0.005, 10, 40, positive
Diplococcus pneumoniae, 0.005, 11, 10, positive
"""
drug_color = OrderedDict([
("Penicillin", "#0d3362"),
("Streptomycin", "#c64737"),
("Neomycin", "black" ),
])
gram_color = OrderedDict([
("negative", "#e69584"),
("positive", "#aeaeb8"),
])
df = pd.read_csv(StringIO(antibiotics),
skiprows=1,
skipinitialspace=True,
engine='python')
width = 800
height = 800
inner_radius = 90
outer_radius = 300 - 10
minr = sqrt(log(.001 * 1E4))
maxr = sqrt(log(1000 * 1E4))
a = (outer_radius - inner_radius) / (minr - maxr)
b = inner_radius - a * maxr
def rad(mic):
return a * np.sqrt(np.log(mic * 1E4)) + b
big_angle = 2.0 * np.pi / (len(df) + 1)
small_angle = big_angle / 7
p = figure(plot_width=width, plot_height=height, title="",
x_axis_type=None, y_axis_type=None,
x_range=(-420, 420), y_range=(-420, 420),
min_border=0, outline_line_color="black",
background_fill_color="#f0e1d2")
p.xgrid.grid_line_color = None
p.ygrid.grid_line_color = None
# annular wedges
angles = np.pi/2 - big_angle/2 - df.index.to_series()*big_angle
colors = [gram_color[gram] for gram in df.gram]
p.annular_wedge(
0, 0, inner_radius, outer_radius, -big_angle+angles, angles, color=colors,
)
# small wedges
p.annular_wedge(0, 0, inner_radius, rad(df.penicillin),
-big_angle+angles+5*small_angle, -big_angle+angles+6*small_angle,
color=drug_color['Penicillin'])
p.annular_wedge(0, 0, inner_radius, rad(df.streptomycin),
-big_angle+angles+3*small_angle, -big_angle+angles+4*small_angle,
color=drug_color['Streptomycin'])
p.annular_wedge(0, 0, inner_radius, rad(df.neomycin),
-big_angle+angles+1*small_angle, -big_angle+angles+2*small_angle,
color=drug_color['Neomycin'])
# circular axes and lables
labels = np.power(10.0, np.arange(-3, 4))
radii = a * np.sqrt(np.log(labels * 1E4)) + b
p.circle(0, 0, radius=radii, fill_color=None, line_color="white")
p.text(0, radii[:-1], [str(r) for r in labels[:-1]],
text_font_size="11px", text_align="center", text_baseline="middle")
# radial axes
p.annular_wedge(0, 0, inner_radius-10, outer_radius+10,
-big_angle+angles, -big_angle+angles, color="black")
# bacteria labels
xr = radii[0]*np.cos(np.array(-big_angle/2 + angles))
yr = radii[0]*np.sin(np.array(-big_angle/2 + angles))
label_angle=np.array(-big_angle/2+angles)
label_angle[label_angle < -np.pi/2] += np.pi # easier to read labels on the left side
p.text(xr, yr, df.bacteria, angle=label_angle,
text_font_size="12px", text_align="center", text_baseline="middle")
# OK, these hand drawn legends are pretty clunky, will be improved in future release
p.circle([-40, -40], [-370, -390], color=list(gram_color.values()), radius=5)
p.text([-30, -30], [-370, -390], text=["Gram-" + gr for gr in gram_color.keys()],
text_font_size="9px", text_align="left", text_baseline="middle")
p.rect([-40, -40, -40], [18, 0, -18], width=30, height=13,
color=list(drug_color.values()))
p.text([-15, -15, -15], [18, 0, -18], text=list(drug_color),
text_font_size="12px", text_align="left", text_baseline="middle")
output_file("burtin.html", title="burtin.py example")
show(p)
其他可视化作品我们直接放出结果,绘制代码省略,大家可自行去官网搜索哈:
periodic
markers
以上所有的可视化作品都是可以交互操作的哦,除此之外,Bokeh 还提供大量的可视化APP应用,具体内容,感兴趣的小伙伴可自行搜索哈~~
总结
这一期我们分享了Python-Bokeh库绘制的可视化作品,体验了Python用于绘制交互式可视化作品放入方便性,还是那句话,适合自己的才是最好的,不要纠结所使用的工具哈,让我们一起探索数据可视化的魅力吧~~
参考来源:https://docs.bokeh.org/en/latest/docs/gallery.html
点分享 点收藏 点点赞 点在看
评论